CLC Server data import and export
This section covers general points related to importing data to and exporting data from a CLC Server when working with the CLC Genomics Workbench. General information about data import and data export is in Importing data and Exporting data and graphics, respectively.
Detailed information about CLC Server configuration is in the server administration manual at: https://resources.qiagenbioinformatics.com/manuals/clcserver/current/admin/index.php?manual=Introduction.html
Data import to a CLC Server
The general location of the data to import is specified from a drop-down list (figure 6.5). If direct data transfer from client systems is enabled on the CLC Server, the option "File system" is included in that list, allowing files on the local system to be selected for import.
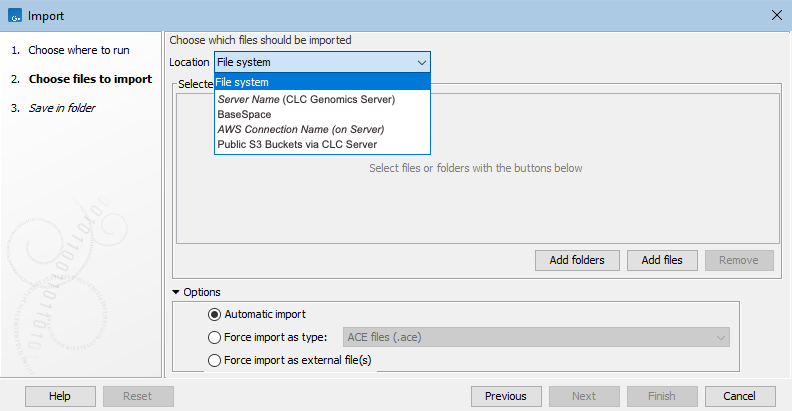
Figure 6.5: When an import is run on a CLC Server, the list of locations that data can imported from reflects the server configuration.
When the option represented in figure 6.5 as "Server Name (CLC Genomics Server)" is selected, the available import/export directories on that CLC Server will be listed in the "Server files" area. When an AWS location is selected, the available S3 buckets will be listed. When BaseSpace is selected, you will be prompted to log into BaseSpace before proceeding.
When "File system" is selected, the CLC Genomics Workbench must maintain its connection to the CLC Server during the first part of the import process, data upload. Further details about this are in Processes tab and Status bar.
Note that when importing data from an AWS S3 bucket, the data is first downloaded from AWS, which AWS charges for.
Data export from a CLC Server
When the CLC Genomics Workbench is connected to a CLC Server, data in CLC Server File System Locations can be exported. The choice of execution environment, as well as settings on the Workbench and Server, affect the locations data can be selected for export from and where the exported files can be saved to:
- Running the export on the Workbench:
- Data in Workbench or Server CLC File System Locations can be selected for export.
- Exported files can be saved to areas the CLC Genomics Workbench has access to, including AWS S3 buckets if an AWS S3 Connection has been configured in the CLC Genomics Workbench.
- Running the export on the CLC Server or Grid via CLC Server:
- Data from Server File System Locations can be can be selected for export.
- Exported files can be saved to Server import/export directories or to an AWS S3 bucket if an AWS Connection has been configured in the CLC Server.
