Create local BLAST databases
You can create a BLAST database from DNA, RNA, or protein sequences to use in local BLAST searches.
The BLAST database files will be saved to an area that is specified as a BLAST database location (see Manage BLAST databases). The BLAST databases found in these locations are listed when launching the BLAST tool (see BLAST against local data).
Making pre-existing BLAST databases available for searching using your Workbench is described in Add pre-formatted BLAST databases.
To create a BLAST database, go to:
Tools | BLAST (![]() )|
Create BLAST Database (
)|
Create BLAST Database (![]() )
)
This opens the dialog seen in figure 16.17.
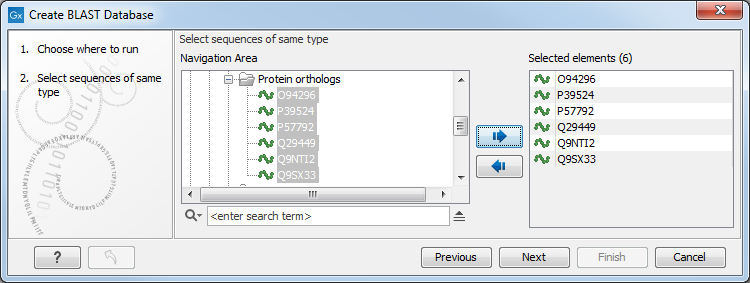
Figure 16.17: Select sequences to be used in the creation of a BLAST database.
After selecting the sequences or sequence lists to include in your database and clicking on Next, you provide information about the BLAST database being made figure 16.18:
- Name. The name of the BLAST database. This name will be used when running BLAST searches and also as the base file name for the BLAST database files.
- Description. A short description. This is displayed along with the database name in the list of available databases when launching a local BLAST search. If no description is entered, the creation date is used as the description.
- Location. The BLAST database location to save the BLAST database files to.
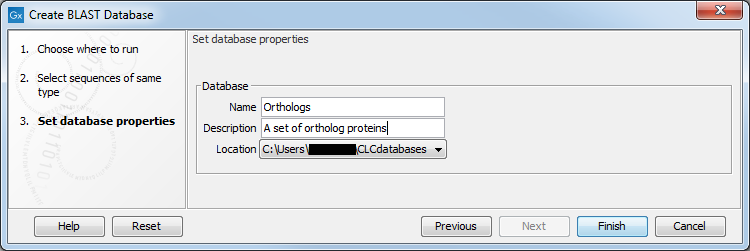
Figure 16.18: Provide information about the BLAST database being created and specify where the files should be saved to.
Click Finish to create the BLAST database. Once the process is complete, the new database will be available in the Manage BLAST databases dialog, and when launching BLAST against local data.
Create BLAST Database creates BLAST+ version 4 (dbV4) databases.
Sequence identifiers and BLAST databases
Restrictions on sequence identifier lengths, format, and duplicates present in the underlying BLAST+ program for making databases, makeblastdb, do not apply when making databases using Create BLAST Database.
Internal handling of sequence names, introduced in version 21.0, allows this level of naming flexibility with newer versions of BLAST+. There should be no obvious effects of this internal handling of sequence names on local BLAST search results, including the names written to BLAST reports. For further details, see the FAQ Can I search BLAST databases made using Create BLAST Databases with BLAST+ software? at https://qiagen.my.salesforce-sites.com/KnowledgeBase/KnowledgeNavigatorPage?id=kA41i000000CjJ9CAK
