Merge Overlapping Pairs
Some paired end library preparation methods using relatively short fragment size will generate data with overlapping pairs. This type of data can be handled as standard paired-end data in the CLC Genomics Workbench. (For variant detection related information, see Detailed information about overlapping paired reads).
In some situations it can be useful to merge each overlapping pair into a single sequence read. This generates longer reads, and can improve the quality, as normally the quality drops towards the end of a read. By overlapping the ends of two reads, the resulting consensus read reflects two read ends instead of just one.
Merge overlapping paired reads by going to:
Tools | Utility Tools (![]() ) | Sequences (
) | Sequences (![]() ) | Merge Overlapping Pairs (
) | Merge Overlapping Pairs (![]() )
)
As input, select one or more sequence lists containing paired reads in forward-reverse orientation.
The resulting merged reads will be in the forward orientation.
Note: Length trimming of reads can be done before or after merging, however the merged read's 3' is the 5' of the reverse read, so that trimming the original reads using the Remove 5' terminal nucleotides option corresponds to trimming the merged reads using both the Remove 5' terminal nucleotides option and the Remove 3' terminal nucleotides option.
The options available in the launch wizard are shown in figure 37.45.
Figure 37.45: Merging overlapping pairs configuration options.
The tool options are best explained in the context of the merging algorithm: Because the fragment size is not an exact number of base pairs and is different from fragment to fragment, an alignment of the two reads has to be performed. If the alignment is good and long enough, the reads will be merged. Good enough in this context means that the alignment has to satisfy some user-specified score criteria (details below). Because of sequencing errors that typically are more abundant towards the end of the read, the alignment is not expected always to be perfect, and the user can decide how many errors are acceptable. Long enough in this context means that the overlap between the reads has to be non-coincidental. Merging two reads that do not really overlap, leads to errors in the downstream analysis, thus it is very important to make sure that the overlap is big enough. If only a few bases overlap was required, some read pairs will match by chance, so this has to be avoided.
The following options define what is good enough and long enough
- Mismatch cost The alignment awards one point for a match, and the mismatch cost is set by this parameter. The default value is 2.
- Gap cost This is the cost for introducing an insertion or deletion in the alignment. The default value is 3.
- Max unaligned end mismatches The alignment is local, which means that a number of bases can be left unaligned. If the quality of the reads is dropping to be very poor towards the end of the read, and the expected overlap is long enough, it makes sense to allow some unaligned bases at the end. However, this should be used with great care which is why the default value is 0. As explained above, a wrong decision to merge the reads leads to errors in the downstream analysis, so it is better to be conservative and accept fewer merged reads in the result.
- Minimum score This is the minimum score of an alignment to be accepted for merging. The default value is 10. As an example: with default settings, this means that an overlap of 13 bases with one mismatch will be accepted (12 matches minus 2 for a mismatch).
Please note that even with the alignment scores above the minimum score specified in the tool setup, the paired reads also need to have the number of end mismatches below the "Maximum unaligned end mismatches" value specified in the tool setup to be qualified for merging.
After configuring the merging options, the option to create a report is offered.
The main result will be two sequence lists for each list in the input: one containing the merged reads (marked as single end reads), and one containing the reads that could not be merged (still marked as paired data). Since the CLC Genomics Workbench handles a mix of paired and unpaired data, both of these sequence lists can be used in the further analysis. However, please note that low quality can be one of the reasons why a pair cannot be merged. Hence, the list of reads that could not be paired is more likely to contain more reads with errors than the one with the merged reads.
Quality scores come into play in two different ways when merging overlapping pairs.
First, if there is a conflict between the reads in a pair (i.e. a mismatch or gap in the alignment), quality scores are used to determine which base the merged read should have at a given position. The base with the highest quality score will be the one used. In the case of gaps, the average of the quality scores of the two surrounding bases will be used. In the case that two conflicting bases have the same quality or both reads have no quality scores, an [IUPAC ambiguity code](see IUPAC codes for nucleotides) representing these bases will be inserted.
Second, the quality scores of the merged read reflect the quality scores of the input reads.
We assume independence of errors in calculating the new quality score for a merged position and carry out the following calculations:
- When the two reads agree at a position, the two quality scores are summed to form the quality score of the base in the new read. The score is capped at the maximum value on the quality score scale which is 64. Phred scores are log scores, so their sums represent the multiplication of the original error probabilities.
- If the two bases disagree at a position, the quality score of the base in the new read is determined by subtracting the lowest score from the highest score of the input reads. Similar to the addition of scores when bases are the same, this adjusts the error probability to reflect a decreased certainty that the base reported at that position is correct.
If a base at a given position in one read of an overlapping region has a very low quality score and the base at that position in the other read has a high score, it is likely that the base with the high quality score is correct. The adjusted quality score for this position in the merged read would reflect that there is less certainty in the base at that position than before. However, such a position would still be assigned quite a high quality, as the base call is still likely to be correct.
Figure 37.46 shows an example of the report generated when merging overlapping pairs.
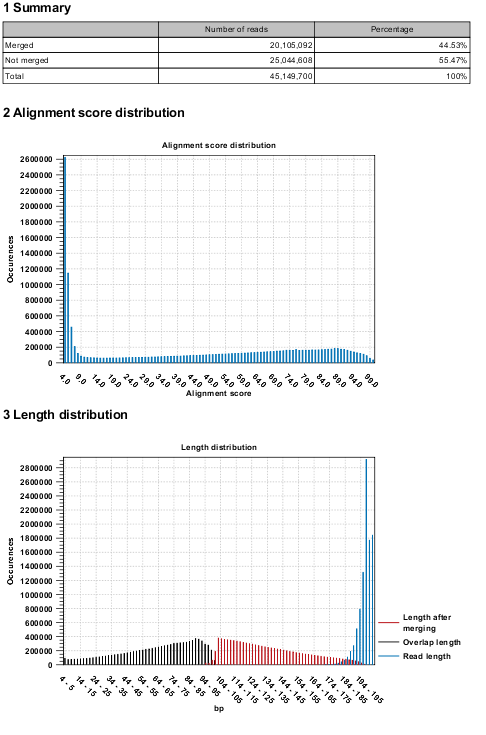
Figure 37.46: Report of overlapping pairs.
It contains three sections:
- A summary showing the numbers and percentages of reads that have been merged.
- A plot of the alignment scores. This can be used to guide the choice of minimum alignment score as explained in Merge overlapping pairs.
- A plot of read lengths. This shows the distribution of read lengths for the pairs that have been merged:
- The length of the overlap.
- The length of the merged read.
- The combined length of the two reads in the pair before merging.
