De Novo Assemble Long Reads parameters
To run the De Novo Assemble Long Reads tool, go to:
Tools | De Novo Sequencing (![]() ) | De Novo Assemble Long Reads (
) | De Novo Assemble Long Reads (![]() )
)
Select one or more sequence lists containing long reads.
Click Next to set the parameters for the assembly (figure 35.28):
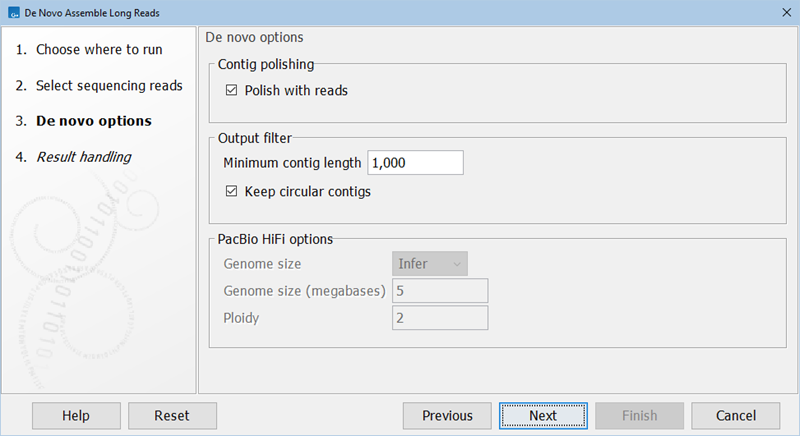
Figure 35.28: The De Novo Assemble Long Reads parameters.
- Oxford Nanopore and PacBio CLR options (enabled for Oxford Nanopore and PacBio CLR reads only)
- Polish with reads. When this parameter is set, two iterations of read polishing are run on the initial contigs. Disabling this parameter results in raw contigs with error rates similar to the error rate of the reads.
- Trim windows (available when Polish with reads is set). When this parameter is set, the assembler will trim the ends of polishing windows according to read coverage during the last polishing round. This can help remove low-quality ends of windows, but may also remove real sequence if the coverage is low.
- Minimum coverage. Sets the minimum read coverage required for a contig to be included in the final contigs list.
- PacBio HiFi options (enabled for PacBio HiFi reads only)
- Genome size. Infer automatically determines the genome size as part of the analysis. Manual instructs the algorithm to use the genome size specified in the text field below for inferring read coverage.
- Genome size (megabases). Enter the expected genome size.
- Ploidy. The number of expected alleles. If it is set to >2, the quality of the assembly for polyploid genomes might be improved.
- Minimum contig length. The minimum length of contigs included in the output. Shorter contigs will be filtered.
- Keep circular contigs. When enabled, the minimum contig length filtering is not applied to circular contigs. This means that all circular contigs will be output regardless of length.
When running running De Novo Assemble Long Reads in a workflow, the workflow element dialog will contain an additional option:
- PacBio HiFi. Check this option to indicate that input reads are PacBio HiFi reads. When selected, the tool will run with an algorithm optimized for HiFi reads.
When running the De Novo Assemble Long Reads tool separately, the read type is inferred from the input reads.
