Searching SRA
Search for Reads in SRA searches the NCBI SRA database for entries with metadata matching the query terms you provide. The sequences and metadata for selected runs can be downloaded. Alternatively, just the metadata for the selected runs can be saved for use later.
Search for Reads in SRA uses NCBI's e-utilities, which occasionally experience downtime. If searches unexpectedly return no results, go to https://www.ncbi.nlm.nih.gov/sra/ to check the status of the service.
Start Search for Reads in SRA by going to:
Download | Search for Reads in SRA (![]() )
)
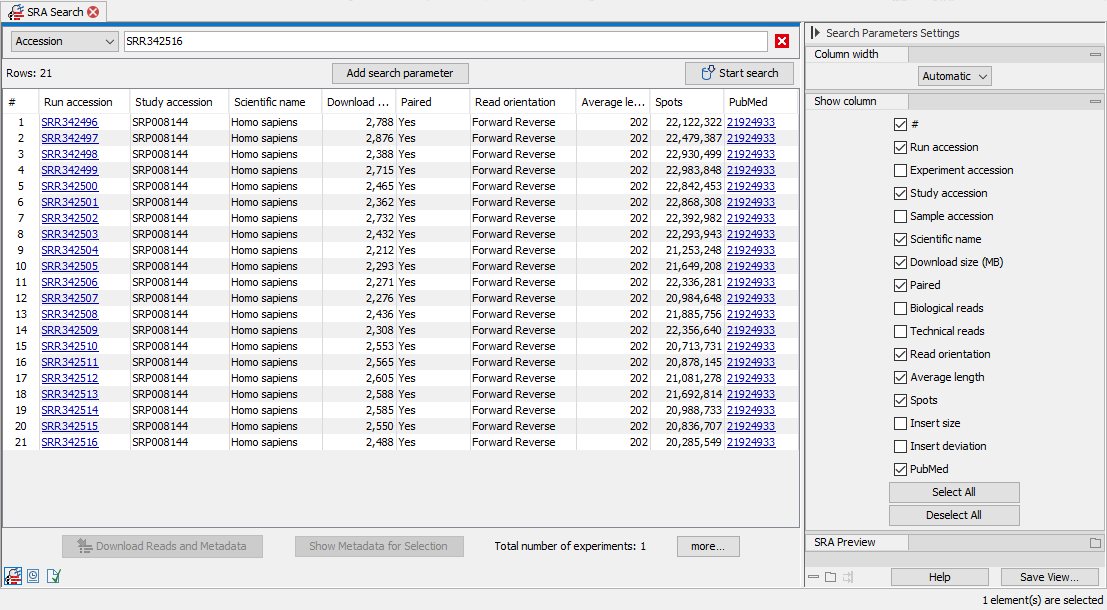
Figure 10.5: "Accession" was selected from the drop-down menu of search fields, and a single accession was entered. The results of the search include the run with the accession provided, as well as runs submitted to SRA as part of the same experiment.
Search fields
A drop-down menu at the top left lists the fields that can be searched (figure 10.5). Generally, all search terms provided must be present in an SRA entry for it to be returned. Within a single search field, OR can be added between terms to indicate that just one of the terms needs to match. The exception to this is the Accession field, which is described further below.
Details about selected search fields:
- All Fields All fields are searched with the terms provided.
Example queries:
"Plasmodium falciparum" "Plasmodium vivax"would yield a list of runs where both terms were found within any of the fields searched."Plasmodium falciparum" OR "Plasmodium vivax"would yield a list of runs where either term was found within any of the fields searched.SRR20016268 SRR20016241 SRR20016341would return no results because an "AND" is assumed between each term. Contrast this with searching in the Accession field with the same terms, described below. - Accession Provide a single accession, or multiple accessions separated by spaces, commas or semicolons. Each term is searched for individually. I.e. terms in a single Accession field are handled as if they were separated by OR. Unlike in other fields writing "OR" between terms is not supported as it is already assumed.
Example query:
SRR20016268 SRR20016241 SRR20016341would return 3 entries.The run with a specified accession is returned, as well as any other runs that were submitted as part of the same experiment. To retrieve all runs in a study, provide the study accession as the query term.
- Modification/Publication date Find entries modified/published within a date range, specified as [MM] [YYYY]. For example, a publication date given the following range
08 2016 to 08 2016would return results published between the first and last days in August 2016. - Strategy Select from a drop-down list of types of experiments e.g., RNA-Seq, ChIP-Seq, etc.
- Library Selection Select from a drop-down list of known library preparation methods, e.g. Poly(A), Size fractionation, etc.
- Platform Select from a drop-down list of NGS sequencing platforms e.g., Illumina, Ion Torrent, etc. Note: Download of data from some platforms, such as Complete Genomics, is not supported.
- Instrument Select from a drop-down list of individual NGS sequencing machines e.g., HiSeq X Ten, Ion Torrent PGM, etc.
- Paired Status The options are Paired and Single. When Paired is selected, SRA runs specified as paired are returned. Selecting Single returns runs where paired status has not been specified.
- Availability Select Public or dbGaP. The latter contains confidential data. Entries in dbGAP can be searched and metadata returned can be saved, but reads cannot be downloaded directly. Access to dbGAP involves an application to the NCBI.
- PubMed Select "has abstract" to find entries with a PubMed abstract or "has full-text article" for entries where the entire publication is available.
SRA search results
A table with one row per result is returned. The columns to display in the table can be configured in the side panel, on the right.
Details about selected column contents:
- Run Accession The accession for an SRA run, hyperlinked to the relevant NCBI webpage, where additional information can be found.
- Download size The size of the SRA format (.sra) file for that run. At least twice this amount of space should be available as temporary space during download and import. See Downloading reads and metadata from SRA for more on space requirements.
- Biological reads The number of biological reads per spot. If there is no read type information for that run in SRA, all reads are assumed to be biological.
- Technical reads The number of technical reads per spot. If there is no read type information for that run in SRA, the value will be 0.
- Read orientation Relevant for paired reads. Unknown means there is no orientation information for that run in SRA. This is always the case for single end reads, but is also frequently the case for paired reads. For such paired end runs, Forward-Reverse orientation is assumed by default when importing, but this is configurable.
- Average length The average length of all the reads in a spot combined. The read lengths in the imported sequence list may differ from these values if you choose not to download all the reads available for the run. E.g. downloading just biological reads when technical reads are also available.
- PubMed If a PubMed entry is associated with the run, it is listed and hyperlinked to the relevant Pubmed webpage.
When a run is selected in the table, the title and abstract for the SRA experiment it is part of is displayed in the SRA Preview tab, under the column configuration section of the side panel.
Please refer to the SRA documentation at the NCBI for full information on the data and metadata available https://www.ncbi.nlm.nih.gov/sra/.
Further details about download and import of data from SRA, including information on file sizes and paired read handling, is provided in Downloading reads and metadata from SRA.
The total number of experiments found is reported at the bottom of the search table. An experiment may have more than one run associated with it.
By default, up to 50 results are retrieved at a time. Click on the more... button below the table to pull additional results, 50 at a time. This number can be configured in Preferences:
Edit | Preferences (![]() ) | General | Number of hits (NCBI/Uniprot)
) | General | Number of hits (NCBI/Uniprot)
Right-click on a row in the results table to get a list of possible additional searches, based on the selected run (figure 10.6).
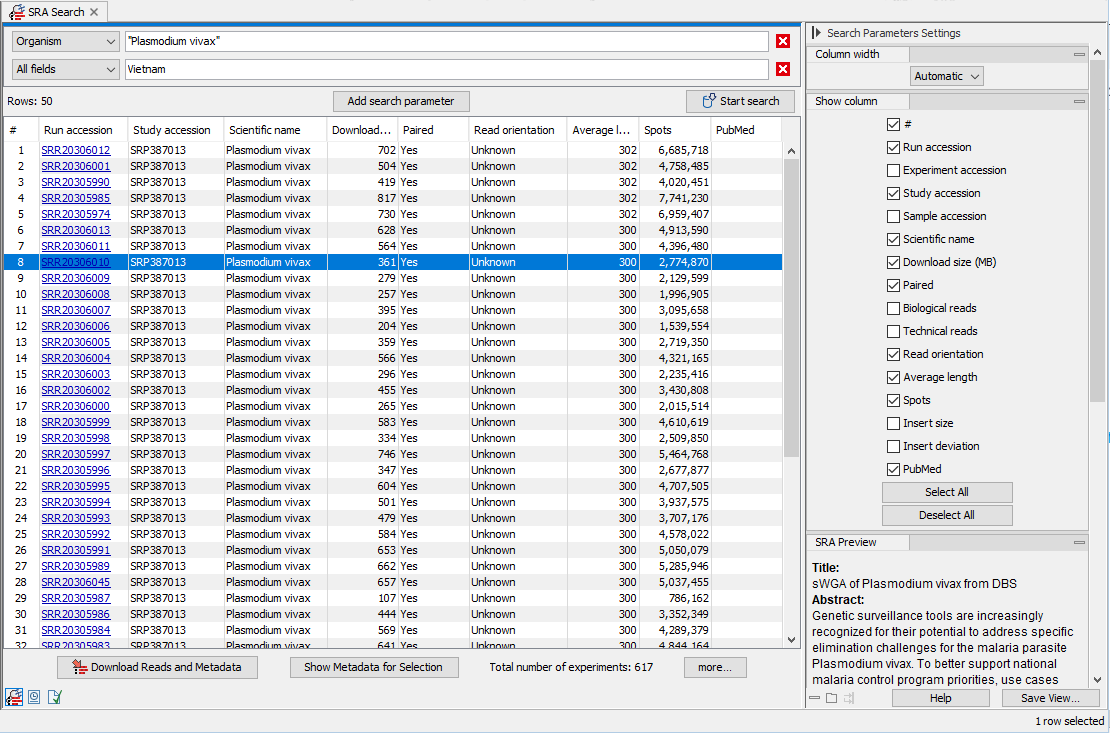
Figure 10.6: The SRA search result table.
