Output from Demultiplex Reads
The outputs from Demultiplex Reads are (see figure 28.24):
- Sequence lists containing the demultiplexed reads, one for each barcode with reads associated with it.
- A sequence list containing the reads without a match to any barcode. The name of this element ends in 'Not grouped'. This output is optional.
- A report summarizing the number of reads identified for each barcode and the number without a match to any barcode ('Not grouped') (figure 28.25). This output is optional.
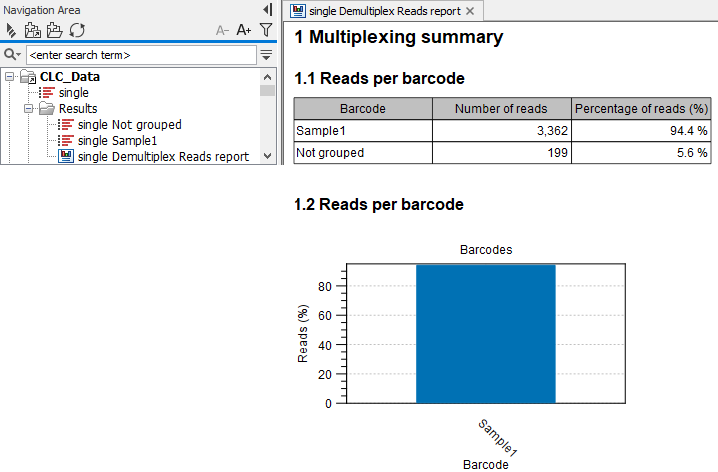
Figure 28.24: Outputs from Demultiplex Reads where the input sequence list named 'single' contained reads matching one barcode CCT named 'Sample1'. Those reads are output in the list named 'single Sample1'. Reads not grouped with any barcode are in the list called 'single Sample1'. The report contains a summary of the number of reads in each of the sequence lists.
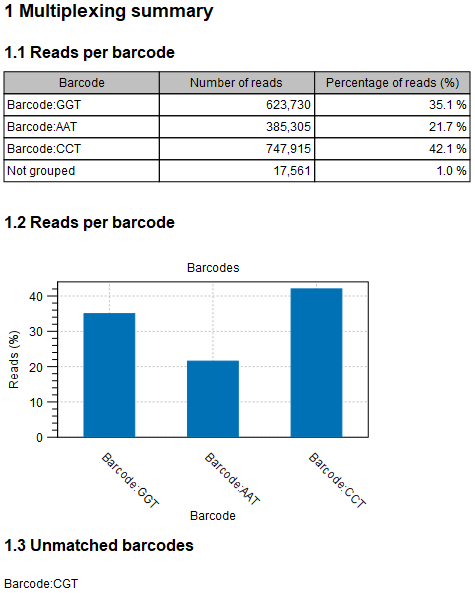
Figure 28.25: An example of a report showing the number of reads in each group. In this example four different barcodes were used to separate four different samples. No reads have been identified for one of the provided barcodes.
There is also an option to create subfolders for each sequence list. This can be useful if downstream analyses will be run in batch mode.
A new sequence list will be generated for each barcode for which reads have been identified, containing all the sequences where this barcode is identified. Both the linker and barcode sequences are removed from each of the sequences in the list, so that only the target sequence remains. This means that you can continue the analysis by doing trimming or mapping. Note that you have to perform separate mappings for each sequence list.
