De Novo Assemble Long Reads and Polish with Short Reads
The De Novo Assemble Long Reads and Polish with Short Reads workflow performs de novo assembly of long reads and polishes the assembly with high quality short reads.
Launching the workflow
The De Novo Assemble Long Reads and Polish with Short Reads workflow is at:
Workflows | Template Workflows | Basic Workflow Designs (![]() )
| De Novo Assemble Long Reads and Polish with Short Reads (
)
| De Novo Assemble Long Reads and Polish with Short Reads (![]() )
)
Launch the workflow and step through the wizard:
- Select the sequence list containing the long uncorrected reads to be assembled (figure 14.97).
- Select the short reads to be used for polishing (figure 14.98).
- Choose assembly parameters. The Minimum contig length can be adjusted to control which contigs are being output. It is particularly important to lower this value if assembling a small microbial genome, as the default may be too high for viral genomes or plasmids. The genome size and ploidy settings become available by checking the PacBio HiFi box. Adjusting these can help improve results when assembling PacBio HiFi reads. (figure 14.99).
- If the short reads have adapters that have not yet been trimmed, a Trim adapter list can be added (figure 14.100).
- Select whether unpolished contigs should be included in the output contig list (figure 14.101).
- In the final step, select a location to save outputs to.
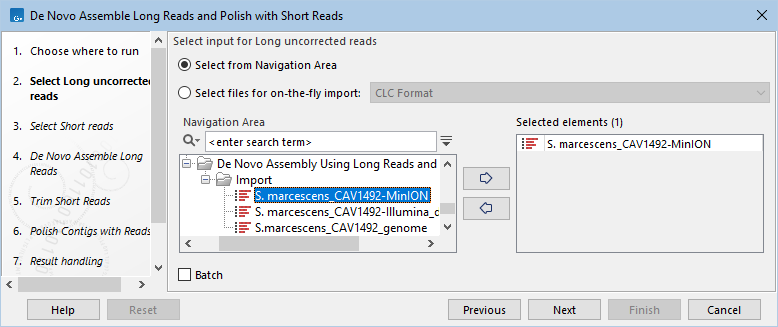
Figure 14.97: Select long uncorrected reads to assemble.
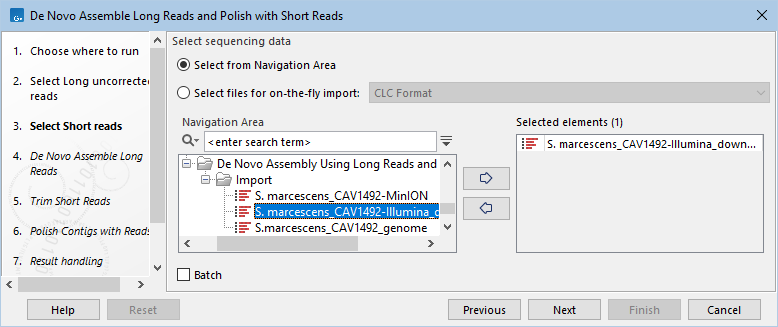
Figure 14.98: Select short reads used to polish the assembly.
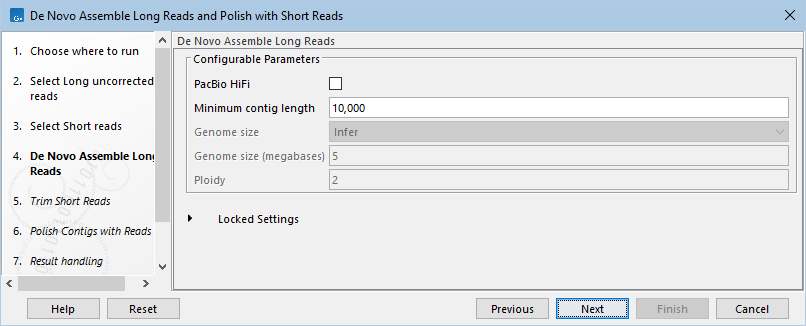
Figure 14.99: Adjust parameters for de novo assembly. The genome size and ploidy settings can only be set if the PacBio HiFi checkbox has been checked. Only do this, if reads are actually PacBio HiFi.
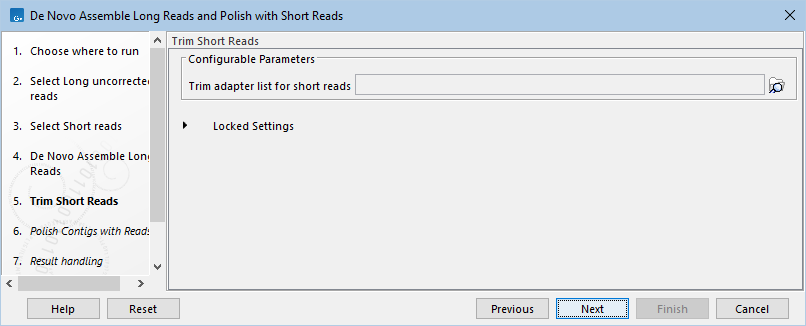
Figure 14.100: Optionally, add a Trim adapter list to use for removing adapters from the short reads.
Figure 14.101: Select whether to keep unpolished contigs.
Tools in the workflow and outputs generated
The De Novo Assemble Long Reads and Polish with Short Reads workflow contains five tools:
- QC for Sequencing Reads. Performs basic QC on the long sequencing reads (QC for Sequencing Reads).
- Trim Reads. Trims the short reads for low quality nucleotides (Trim Reads).
- De Novo Assemble Long Reads. Performs de novo assembly of the long input reads (De Novo Assemble Long Reads). The output contig list is used as input for the polishing step.
- Polish Contigs with Reads. Cleans up errors in the de novo assembled contigs using short, high quality reads (Polish Contigs with Reads). Outputs a sequence list containing the updated contigs.
- Create Sample Report. Collects reports from the individual tools into a single consolidated sample report (Create Sample Report). All stand-alone reports are also output in a "QC & Reports" folder.
Customizing the workflow
Template workflows can be easily edited to add, or change analysis steps. See Template workflows for information about how to open a copy of a template workflow for editing.
- Trim Reads. Other than changing the default quality and optional adapter trimming, additional trimming options can be set to process the short reads before being used for polishing.
- Polish Contigs with Reads. Partial order alignment window size can be adjusted to optimize polishing results.
