Flags
The use of alignment flags by the CLC Genomics Workbench is shown in the following table and subsequent examples.
| Bit | SAM description | Usage |
| 0x1 | Template having multiple segments in sequencing | Set if the segment is part of a pair |
| 0x2 | Each segment properly aligned according to the aligner | Set if the pair is not broken |
| 0x4 | Segment unmapped | Never set since the exporter does not export unmapped reads |
| 0x8 | Next segment in the template unmapped | Never set by the exporter. If a segment has an unmapped mate, the flag 0x1 is not set for the segment, i.e. it is not output as part of a pair |
| 0x10 | SEQ being reverse complemented | Set if and only if the segment was reverse complemented during mapping |
| 0x20 | SEQ of the next segment in the template being reversed | Set if and only if the mate was reverse complemented during mapping |
| 0x40 | The first segment in the template | This mate is the first segment of the pair |
| 0x80 | The last segment in the template | This mate is the second segment of the pair |
| 0x100 | Secondary alignment | Never set by the exporter. No reads with this flag set are imported. |
| 0x200 | Not passing quality controls | Never set by the exporter and ignored by the importer |
| 0x400 | PCR or optical duplicate | Never set by the exporter and ignored by the importer |
Flag Examples
The following table illustrates some of the possible flags in the CLC Genomics Workbench.
| Description of the example | Bits | Flag | Illustration |
| The first mate of a non-broken paired read | 0x1, 0x2, 0x20, 0x40 | 99 | Figure I.1 |
| The second mate of a non-broken paired read | 0x1, 0x2, 0x10, 0x80 | 147 | Figure I.2 |
| A single, forward read (or paired read, where only one mate of the pair is mapped) | No set bits | 0 | Figure I.3 |
| A single, reversed read (or paired read, where only one mate of the pair is mapped) | 0x10 | 16 | Figure I.4 |
| The first, forward segment from a broken pair with forward mate | 0x1, 0x40 | 65 | Figure I.5 |
| The second, forward segment from broken pair with reversed mate | 0x1, 0x20, 0x80 | 161 | Figure I.6 |
| The first, reversed segment from broken pair with forward mate | 0x1, 0x10, 0x40 | 81 | Figure I.7 |
| The second, reversed segment from broken pair with reversed mate | 0x1, 0x10, 0x20, 0x80 | 177 | Figure I.8 |
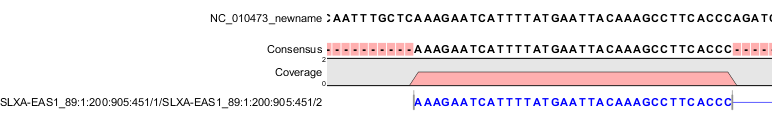
Figure 39.2: The read is paired, both reads are mapped and the mate of this read is reversed
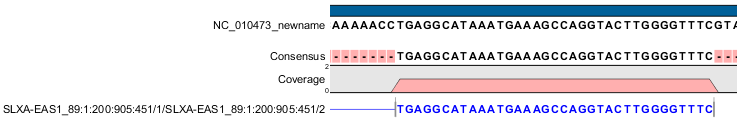
Figure 39.3: The read is paired, both mates are mapped, and this segment is reversed
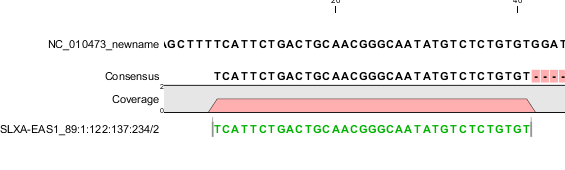
Figure 39.4: A single, forward read, or a paired read where the mate is not mapped
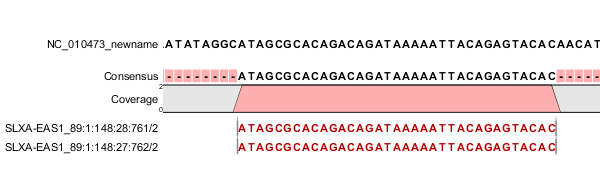
Figure 39.5: The read is a single, reversed read, or a paired read where the mate is not mapped
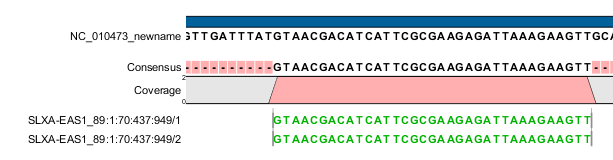
Figure 39.6: These forward reads are paired. They map to the same place, so the pair is broken

Figure 39.7: Forward read that is part of a broken read where the mate is reversed

Figure 39.8: Reversed read that is part of a broken pair, where the mate is forward
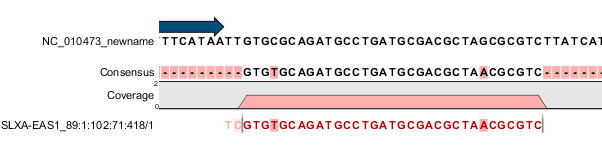
Figure 39.9: Reversed read that is part of a broken pair, where the mate is also reversed.
