Correct Long Reads
This tool has been deprecated and will be retired in a future version of the software. It has been moved to the Legacy Tools (![]() ) folder under the Tools menu, and its name has "(legacy)" appended to it.
) folder under the Tools menu, and its name has "(legacy)" appended to it.
The Correct Long Reads tool was originally designed to enable the correction of error-prone long reads. Over the past years, the quality of long reads has improved dramatically, to the point where correction is no longer needed. In addition, the most common applications such as Map Long Reads to Reference and De Novo Assemble Long Reads can now handle the older error-prone reads without prior correction. Therefore, the tool will be retired.
If you have concerns about the future retirement of this tool, please contact QIAGEN Bioinformatics Support team at ts-bioinformatics@qiagen.com.
The Correct Long Reads tool enables the correction of a set of error-prone long reads by finding overlaps between the reads, and performing a consensus error correction using Racon [Vaser et al., 2017].
Note that if the aim is to create an assembly from a set of error-prone long reads it is not recommended to run Correct Long Reads prior to De Novo Assemble Long Reads. For other applications, however, it may be beneficial to correct the reads before further analysis.
To run the Correct Long Reads tool, go to:
Tools | Legacy Tools | Correct Long Reads (legacy) (![]() )
)
Select one or more sequence lists containing reads.
In the next dialog, set the following parameters (figure 38.1):
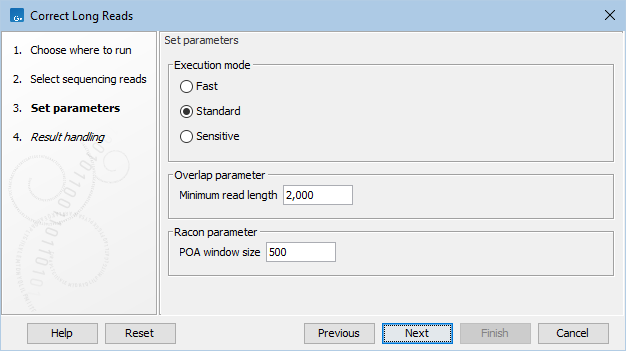
Figure 38.1: Correct Long Reads parameters
- Execution mode.
- Fast. An overlap mapper computes overlaps using a coarse all-to-all sequence alignment, prior to running Racon. This option will use the least amount of memory.
- Standard. Similar to the Fast , but with a higher sensitivity.
- Sensitive. Minimap2 is used to compute a more precise all-to-all sequence alignment prior to running Racon. This option should only be used for low-coverage datasets.
- Minimum read length. Reads shorter than this value will not be included in the correction process.
- POA window size. The window size for which Racon computes partial order alignments (POA). A larger window size enhances the ability to capture more global structure during the polishing process, but it also increases the memory requirement.
Subsections
