AWS Connections
AWS connections are used when:
- Accessing S3 buckets in AWS accounts you have access to, to import data from or export data to.
- Submitting analyses to a CLC Genomics Cloud setup, if available on that AWS account.
Working with data in AWS S3 buckets via the Workbench is of particular relevance when submitting jobs to run on a CLC Genomics Cloud setup making use of functionality provided by the CLC Cloud Module.
When launching workflows to run locally using on-the-fly import and selecting files from AWS S3, the files selected are first downloaded to a temporary folder and are subsequently imported.
All traffic to and from AWS is encrypted using a minimum of TLS version 1.2.
Configuring access to AWS resources
To configure an AWS Connection or manage existing connections, go to:
Connections | AWS Connections (![]() )
)
This opens the Manage AWS Connections dialog (figure 6.6). Existing AWS Connections and their status will be listed. These can be edited or removed. Clicking on the Add AWS Connection button allows a new connection to be configured, as described below.
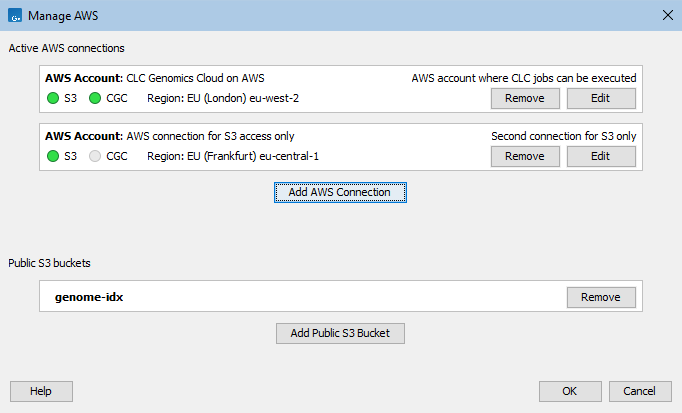
Figure 6.6: The Manage AWS Connections dialog
Configuring an AWS Connection
To configure a new AWS Connection, click on the Add AWS Connection button in the Manage AWS Connections dialog and enter the relevant details for the AWS Connection (figure 6.7). These are:
- Connection name: A short name of your choice, identifying the AWS account. This name will be shown as the name of the data location when importing data to or exporting data from Amazon S3.
- Description: A description of the AWS account (optional).
- AWS access key ID: The access key ID for programmatic access for your AWS IAM user.
- AWS secret access key: The secret access key for programmatic access for your AWS IAM user.
- AWS region: An AWS region. Select from the drop-down list.
- AWS partition: The AWS partition for your account.
The dialog continuously validates the settings entered. When they are valid, the Status box will contain the text "Valid" and a green icon will be shown. Click on OK to save the settings.
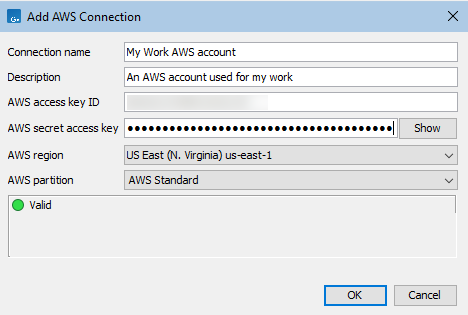
Figure 6.7: Configuration of an AWS Connection in a CLC Workbench
AWS connection status is indicated using colors. Green indicates the connection is valid and ready for use. Connections to a CLC Genomics Cloud are indicated in the CGC column (figure 6.6). To submit analyses to the CLC Genomics Cloud, the CLC Cloud Module must be installed and a license for that module must be available.
AWS credentials entered are stored, obfuscated, in Workbench user configuration files.
Note: Multiple AWS Connections using credentials for the same AWS account cannot be configured.
Configuring access to public S3 buckets is described in Public S3 buckets.
Importing data from AWS S3
AWS S3 buckets for each AWS Connection are available will be listed as possible sources of data in the launch wizards of relevant import tools and workflows when using on-the-fly import (see Launching workflows individually and in batches).
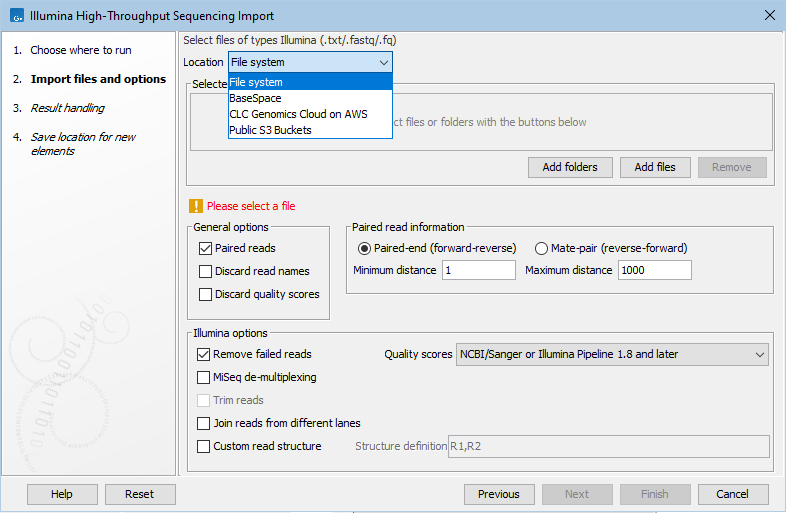
Figure 6.8: Files in local or remote locations can be selected for import by the Illumina importer of the CLC Genomics Workbench.
AWS S3 buckets can be browsed under the Remote Files tab next to the Navigation Area tab, in the top left side of the Workbench (see Working with AWS S3 using the Remote Files tab).
Exporting data to AWS S3
To export data to an AWS S3 bucket, launch the exporter, and when prompted for an export location, select the relevant option from the drop-down menu (figure 6.9).
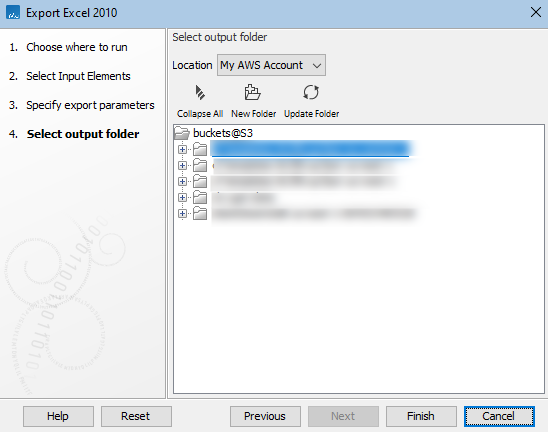
Figure 6.9: After an AWS connection is selected when exporting, you can select the S3 bucket and location within that bucket to export to.
