Find binding sites and create fragments
In CLC Genomics Workbench you have the possibility of matching known primers against one or more DNA sequences or a list of DNA sequences. This can be applied to test whether a primer used in a previous experiment is applicable to amplify a homologous region in another species, or to test for potential mispriming. This functionality can also be used to extract the resulting PCR product when two primers are matched. This is particularly useful if your primers have extensions in the 5' end. Note that this tool is not meant to analyze rapidly high-throughput data. The maximum amount of sequences the tool will handle in a reasonable amount of time depends on your computer processing capabilities.
To search for primer binding sites:
Tools | Molecular Biology Tools (![]() ) | Primers and Probes (
) | Primers and Probes (![]() )| Find Binding Sites and Create Fragments (
)| Find Binding Sites and Create Fragments (![]() )
)
If a sequence was already selected in the Navigation Area, this sequence is now listed in the Selected Elements window of the dialog. Use the arrows to add or remove sequences or sequence lists from the selected elements.
Click Next when all the sequence have been added.
Note! You should not add the primer sequences at this step.
Parameters
This opens the dialog displayed in figure 21.18.
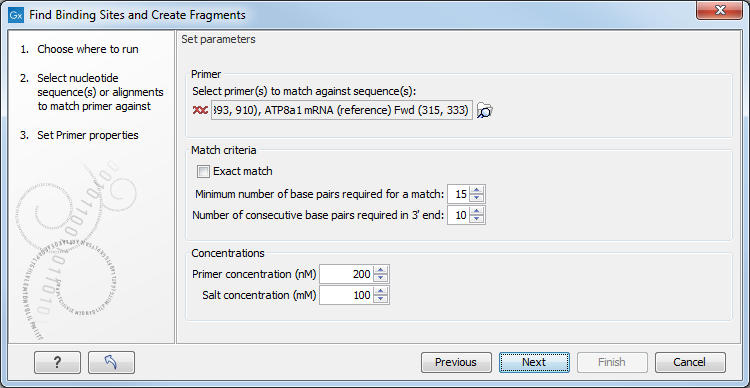
Figure 21.18: Search parameters for finding primer binding sites.
At the top, select one or more primers by clicking the browse (![]() ) button. In CLC Genomics Workbench, primers are just DNA sequences like any other, but there is a filter on the length of the sequence. Only sequences up to 400 bp can be added.
) button. In CLC Genomics Workbench, primers are just DNA sequences like any other, but there is a filter on the length of the sequence. Only sequences up to 400 bp can be added.
The Match criteria for matching a primer to a sequence are:
- Exact match. Choose only to consider exact matches of the primer, i.e. all positions must base pair with the template.
- Minimum number of base pairs required for a match. How many nucleotides of the primer that must base pair to the sequence in order to cause priming/mispriming.
- Number of consecutive base pairs required in 3' end. How many consecutive 3' end base pairs in the primer that MUST be present for priming/mispriming to occur. This option is included since 3' terminal base pairs are known to be essential for priming to occur.
Note that the number of mismatches is reported in the output, so you will be able to filter on this afterwards (see Results - binding sites and fragments).
Below the match settings, you can adjust Concentrations concerning the reaction mixture. This is used when reporting melting temperatures for the primers.
- Primer concentration. Specifies the concentration of primers and probes in units of nanomoles (
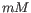 )
)
- Salt concentration. Specifies the concentration of monovalent cations (
![$ [Na^+]$](img40.gif) ,
, ![$ [K^+]$](img41.gif) and equivalents) in units of millimoles (
and equivalents) in units of millimoles (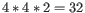 )
)
Subsections
