Advanced preferences
Proxy Settings
CLC Genomics Workbench supports the use of an HTTP-proxy and an anonymous SOCKS-proxy.
You have the choice between an HTTP-proxy and a SOCKS-proxy. The CLC Workbench only supports the use of a SOCKS-proxy that does not require authorization.
You can select whether the proxy should also be used for FTP and HTTPS connections.
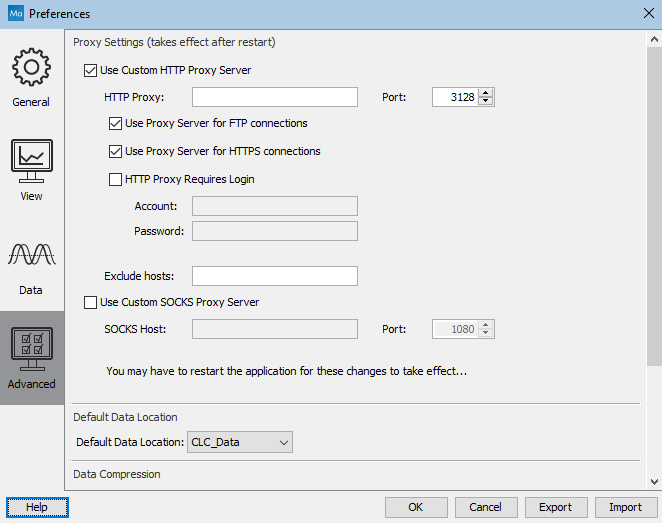
Figure 4.7: Advanced preference settings include proxy configuration options.
List hosts that should be contacted directly, i.e. not via the proxy server, in the Exclude hosts field. The value can be a list, with each host separated by a | symbol. The wildcard character * can also be used. For example: *.foo.com|localhost.
The proxy can be bypassed when connecting to a CLC Server by checking the box next to Bypass proxy when connecting to a CLC Server.
Workbenches can be preconfigured to bypass the proxy settings when connecting to a CLC Server by configuring this setting in a proxy.properties file, where the IP address (not the host name) of the CLC Server is provided in the proxyexclude field. See https://resources.qiagenbioinformatics.com/manuals/workbenchdeployment/current/index.php?manual=Per_computer_Workbench_information.html for further details.
If you have problems with your proxy settings, please contact your systems administrator.
Default data location
The default location is used when you import a file without selecting a folder or element in the Navigation Area first. It is set to the folder called CLC_Data in the Navigation Area, but can be changed to another data location using a drop down list of data locations already added (see Adding locations). Note that the default location cannot be removed, but only changed to another location.
Data Compression
CLC format data is stored in an internally compressed format. The application of internal compression can be disabled by unchecking the option "Save CLC data elements in a compressed format". This option is enabled by default. Turning this option off means that data created may be larger than it otherwise would be.
Enabling data compression may impose a performance penalty depending on the characteristics of the hardware used. However, this penalty is typically small, and we generally recommend that this option remains enabled. Turning this option off is likely to be of interest only at sites running a mix of older and newer CLC software, where the same data is accessed by different versions of the software.
Compatibility information:
- A new compression method was introduced with version 22.0 of the CLC Genomics Workbench, CLC Main Workbench and CLC Genomics Server. Compressed data created using those versions can be read by version 21.0.5 and above, but not earlier versions.
- Internal compression of CLC data was introduced in CLC Genomics Workbench 12.0, CLC Main Workbench 8.1 and CLC Genomics Server 11.0. Compressed data created using these versions is not compatible with older versions of the software. Data created using these versions can be opened by later versions of the software, including versions 22.0 and above.
To share specific data sets for use with software versions that do not support the compression applied by default, we recommend exporting the data to CLC or zip format and turning on the export option "Maximize compatibility with older CLC products". See Export of folders and data elements in CLC format.
NCBI Integration
Without an API key, access to NCBI from asingle IP-address is limited to 3 requests per second; if many workbenches use the same IP address when running the Search for Reads in SRA..., Search for Sequences at NCBI and Search for PDB Structures at NCBI tools they may hit this limit. In this case, you can create an API key for NCBI E-utilities in your NCBI account and enter it here.
NCBI BLAST
The standard URL for the BLAST server at NCBI is: https://blast.ncbi.nlm.nih.gov/Blast.cgi, but it is possible to specify an alternate server URL to use for BLAST searches. Be careful to specify a valid URL, otherwise BLAST will not work.
Read Mapper
It is possible to change the size (in MB) of the Read Mapper reference cache.
BaseSpace configuration
Import from BaseSpace requires no configuration. The settings in this area are optional.
- Illumina regional instance ID Leave this empty to use the default BaseSpace region (USA). Mouse over the field to see the supported regional instances.
- Client ID and Client secret Credentials for a BaseSpace app can be entered here.
Reference Data
URL to use: Reference data sets available under the QIAGEN Sets tab of the Reference Data Manager are downloaded from the URL provided here. In most cases, this setting should not be changed.
Download to CLC Server via: Relevant when the "On Server" option is chosen in the Reference Data Manager, so that data in the CLC_References area of a CLC Genomics Server is used. With the "CLC Server" option chosen (figure 4.8), data downloaded to a CLC Genomics Server using the Reference Data Manager is downloaded directly to the Server, without going via the Workbench.
If the CLC Genomics Server has no access to the external network, but the CLC Workbench does, choose the "CLC Workbench" option. Data is then downloaded to the Workbench and then moved to the reference data area on the Server. The Workbench must be left running throughout the data download process when this option setting is selected.
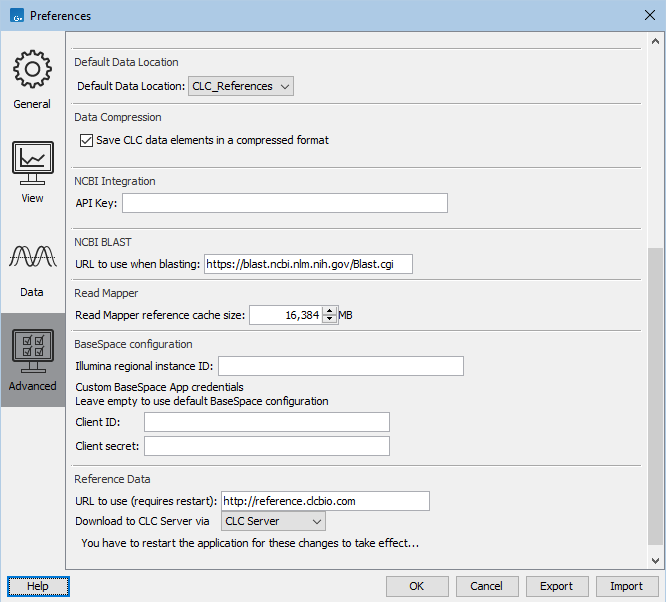
Figure 4.8: Some of the options in the Advanced area of the Workbench Preferences.
