Predict Splice Site Effect
Predict Splice Site Effect takes as input a variant track, and outputs variants annotated to indicate whether they may affect splicing.
To run the tool, go to:
Tools | Resequencing Analysis (![]() ) | Functional Consequences (
) | Functional Consequences (![]() ) | Predict Splice Site Effect (
) | Predict Splice Site Effect (![]() )
)
The following options can be configured (figure 32.21):
- mRNA track An annotation track containing transcript annotations, used to identify splice sites as intron-exon boundaries.
- Splice site window size The number of base pairs around an intron-exon boundary where variants are annotated as causing possible splice site disruption.
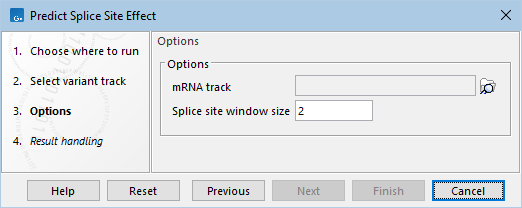
Figure 32.21: Options for Predict Splice Site Effect.
The output variant track contains a new column "Possible splice effect", indicating whether variants may affect splicing (figure 32.22).
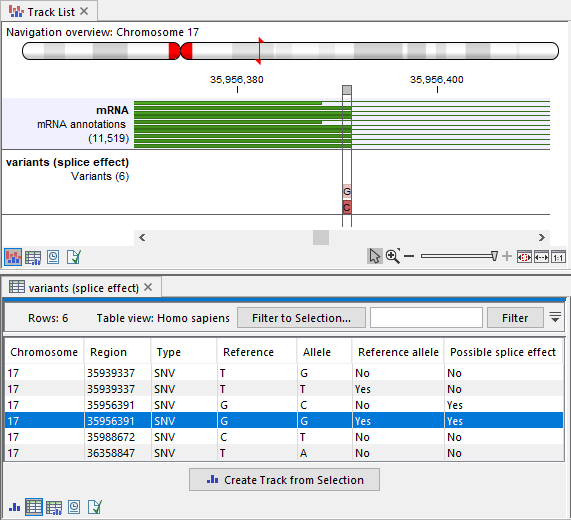
Figure 32.22: Top: A track list displaying an mRNA track along with the output from Predict Splice Site Effect. Bottom: The table view of the output. Each variant includes an additional column, "Possible splice effect", which indicates whether it may impact splicing.
