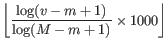BED export
Annotation and expression tracks can be exported to a BED format file using the dedicated BED exporter. See http://genome.ucsc.edu/FAQ/FAQformat.html#format1 for format details. Note that:
- White spaces in the annotation/gene/transcript name are replaced with _.
- The thickStart and thickEnd BED fields are populated with the same values as the chromStart and chromEnd BED fields, respectively. I.e. the start and end positions of the feature in the chromosome or scaffold.
- The itemRgb BELD field is assigned the value 0.
- The score BED field is assigned values as follows:
- For annotation tracks, if the annotation does not contain a score, the value is set to 0. Otherwise, the annotation score is used, where negative scores are set to 0 and scores larger than 1000 are set to 1000.
- For expression tracks, the score reflects the expression value and it is set to
where v is the expression value, and m/M is the lowest/largest observed expression values in the entire expression track.