References and masking
When the sequences are selected, click Next, and you will see the dialog shown in figure 30.2.
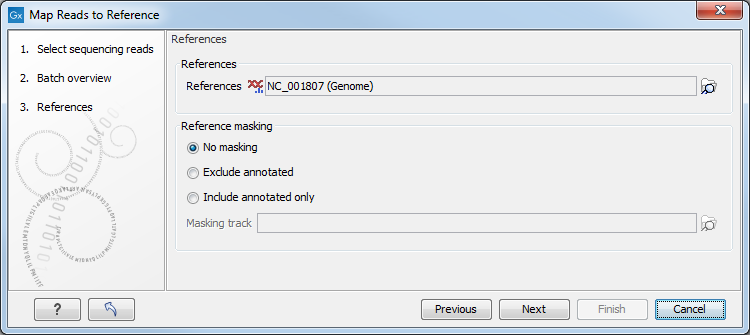
Figure 30.2: Specifying the reference sequences and masking.
At the top, select one or more reference sequences by clicking the
Browse (![]() ) button. You can
select either single sequences, a list of sequences or a sequence
track as reference.
Note the following constraints:
) button. You can
select either single sequences, a list of sequences or a sequence
track as reference.
Note the following constraints:
- single reference sequences longer than 2gb (
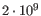 bases) are not supported.
bases) are not supported.
- a maximum of 120 input items (sequence lists or sequence elements) can be used as input to a single read mapping run.
Including or excluding regions (masking)
The next part of the dialog shown in figure 30.2 lets you mask the reference sequences. Masking refers to a mechanism where parts of the reference sequence are not considered in the mapping. This can be useful for example when mapping data is captured from specific regions (e.g. for amplicon resequencing). The output will still include the full reference sequence, but no reads will be mapped in the ignored regions.
Note that you should be careful that your data is indeed only sequenced from the target regions. If not, some of the reads that would have matched a masked-out region perfectly may be placed wrongly at another position with a less-perfect match and lead to wrong results for subsequent variant calling. For resequencing purposes, we recommend testing whether masking is appropriate by running the same data set through two rounds of read mapping and variant calling: one with masking and one without. At the end, comparing the results will reveal if any off-target sequences cause problems in the variant calling.
Masking out repeats or using other masks with many regions is not recommended. Repeats are handled well without masking and do not cause any slowdown. On the contrary, masking repeats is likely to cause a dramatic slowdown in speed, increase memory requirements and lead to incorrect read placement.
To mask a reference sequence, first click the Include or Exclude options, and then click the Browse (![]() ) button to select a track to use for masking.
If you have annotations on a sequence instead of a track, you can convert the annotation type to a track (see Converting data to tracks and back).
) button to select a track to use for masking.
If you have annotations on a sequence instead of a track, you can convert the annotation type to a track (see Converting data to tracks and back).
