Alignment-based TaqMan probe design
CLC Genomics Workbench allows the user to design solutions for TaqMan quantitative PCR which consist of four oligos: a general primer pair which will amplify all sequences in the alignment, a specific TaqMan probe which will match the group of included sequences but not match the excluded sequences and a specific TaqMan probe which will match the group of excluded sequences but not match the included sequences. The selection boxes are used to indicate the status of a sequence, if the box is checked the sequence belongs to the included sequences, if not, it belongs to the excluded sequences. We use the terms included and excluded here to be consistent with the previous section although a probe solution is presented for both groups.
When in TaqMan mode, primers are not allowed degeneracy or mismatches to any template sequence in the alignment, variation is only allowed/required in the TaqMan probes.
Clicking on the Calculate button will prompt the dialog shown in figure 21.15.
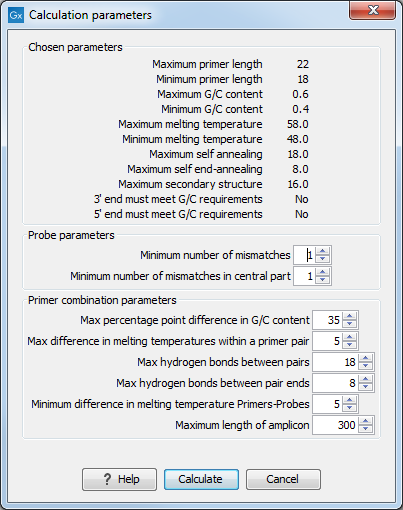
Figure 21.15: Calculation dialog shown when designing alignment-based TaqMan probes.
The top part of this dialog is identical to the Standard PCR dialog for designing primer pairs (figure 21.14).
The central part of the dialog contains parameters to define the specificity of TaqMan probes. Two parameters can be set:
- Minimum number of mismatches. The minimum total number of mismatches that must exist between a specific TaqMan probe and all sequences which belong to the group not recognized by the probe.
- Minimum number of mismatches in central part. The minimum number of mismatches in the central part of the oligo that must exist between a specific TaqMan probe and all sequences which belong to the group not recognized by the probe.
The lower part of the dialog contains parameters pertaining to primer pairs and the comparison between the outer oligos (primers) and the inner oligos (TaqMan probes). Here, six options can be set:
- Max percentage point difference in G/C content. If this is set at e.g. 5 points, a pair of primers with 45% and 50% G/C nucleotides, respectively, will be allowed, whereas a pair of primers with 45% and 51% G/C nucleotides, respectively, will not be included.
- Max difference in melting temperatures within a primer pair. The difference in melting temperatures (in Celsius) that primers in a pair are allowed to have.
- Max hydrogen bonds between pairs. The maximum number of hydrogen bonds allowed between the forward and reverse primer in a pair. This criterion is applied to all possible combinations of primers and probes.
- Max hydrogen bonds between pair ends. The maximum number of hydrogen bonds allowed in the consecutive ends of the forward and reverse primer in a pair.
- Minimum difference in melting temperature Inner-Outer. All comparisons between the melting temperature of primers (outer) and probes (inner) must be at least this different, otherwise the solution set is excluded.
- Maximum length of amplicon. The maximum length of the PCR fragment.
The output of the design process is a table of solution sets. Each solution set contains the following:
- A set of primers which are general to all sequences in the alignment.
- A TaqMan probe which is specific to the set of included sequences (sequences where selection boxes are checked).
- A TaqMan probe which is specific to the set of excluded sequences (marked by *).
Otherwise, the table is similar to that described for TaqMan probe prediction on single sequences.
