Running remove duplicate mapped reads
To run the Remove Duplicate Mapped Reads tool, go to:
Tools | Resequencing Analysis (![]() ) | Remove Duplicate Mapped Reads (
) | Remove Duplicate Mapped Reads (![]() )
)
This opens a dialog where you can select mapping results in reads tracks (![]() ) format. Clicking Next allows you to set the threshold parameters as displayed in figure 30.45.
) format. Clicking Next allows you to set the threshold parameters as displayed in figure 30.45.
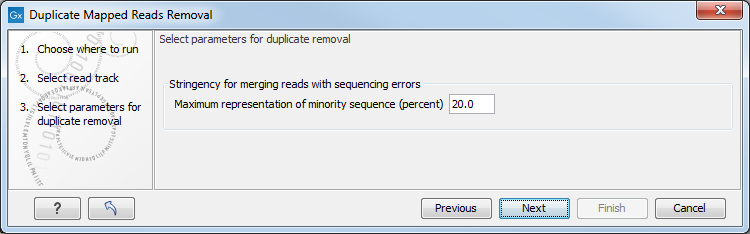
Figure 30.45: Setting the stringency for merging similar reads.
The parameter is explained in detail in Algorithm details and parameters.
Clicking Next will reveal the output options. The main output is a list of the reads that remain after the duplicates have been removed. In addition, you can get the following output:
- List of duplicate sequences
- These are the sequences that have been removed.
- Report
- This is a brief summary report with the number of reads that have been removed (see an example in figure 30.46).
Note! The Remove Duplicate Mapped Reads tool may run this before or after local realignment. The order in which these two tools are run should make little if any difference.
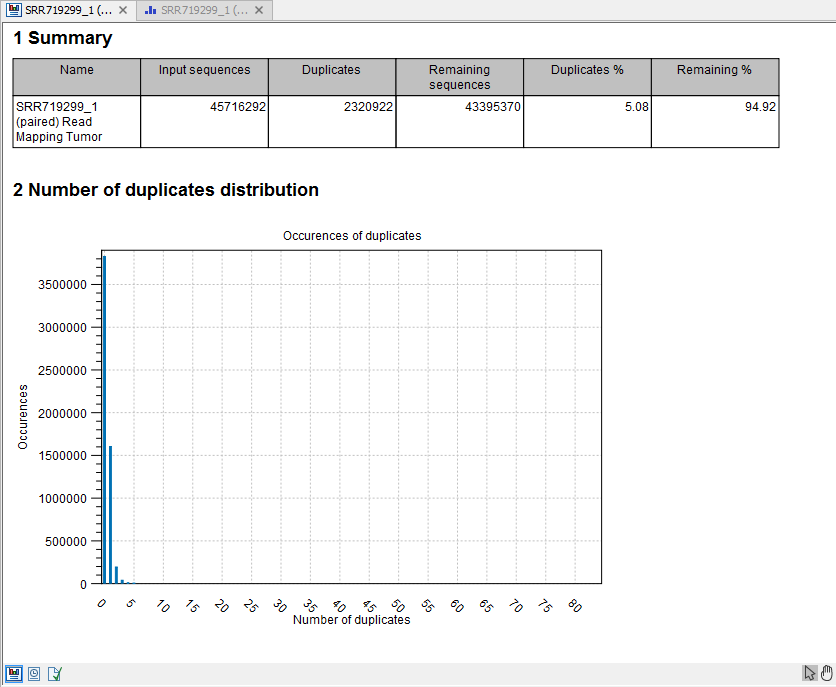
Figure 30.46: Summary statistics on the duplicate mapped reads.
