Ultima Genomics
The Ultima importer can be used for importing CRAM files generated by Ultima sequencing technology.
To launch the Ultima importer, go to:
Import (![]() ) | Other NGS Reads (
) | Other NGS Reads (![]() ) | Ultima (
) | Ultima (![]() ).
).
This opens a dialog where files can be selected and import options specified (figure 7.17).
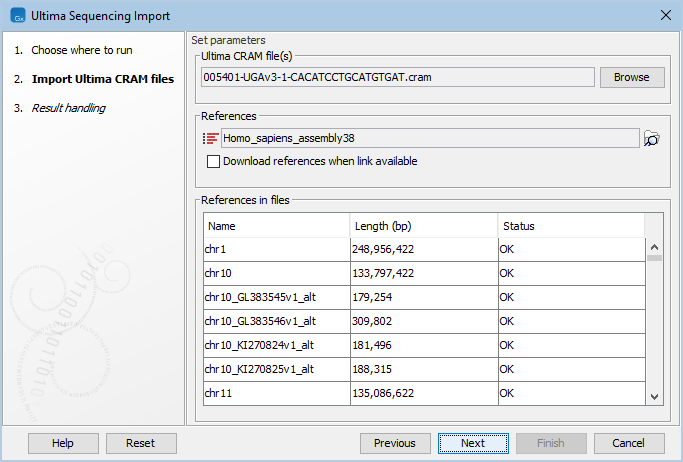
Figure 7.17: Choosing Ultima CRAM file and reference(s).
Providing references for the Ultima CRAM file
The reference sequence(s) that are referred to within the Ultima CRAM file must be specified in the 'Set parameters' wizard step (figure 7.17):
- If the Ultima CRAM file already contains information about where to find the reference(s), tick Download references when link available.
- If the input reference(s) are present in the CLC Genomics Workbench, click on the "Find in folder" icon (
 ) to select the reference(s).
) to select the reference(s).
Occurrences of disallowed characters according to the specification at https://samtools.github.io/hts-specs/SAMv1.pdf (whitespaces \ , " ` ' @ ( ) [ ] < >) in the input references are replaced by _ (underscore). Additionally, = and * are only disallowed at the beginning of the reference names. E.g., an input reference named
*my=reference@sequenceis considered the same as the reference_my=reference_sequencereferred to within the Ultima CRAM file.
The table under 'References in files' contains the references that are referred to within the Ultima CRAM file, with their name, length, and a status. The status indicates whether a given reference referred to within the Ultima CRAM file is present in the input references. The status can be:
- OK. There is a reference in the input references with this name and length.
- Length differs. There is a reference in the input references with this name, but with a different length.
- Download link available. The Ultima CRAM file contains a URL for this reference. Tick Download references when link available to automatically download the reference.
- Will download. The Ultima CRAM file contains a URL for this reference and Download references when link available is already ticked. The reference is automatically downloaded.
- Missing, download link not available. There is no reference in the input references with this name, and there is no URL available in the Ultima CRAM file for downloading the reference.
A reference is 'matched' when the status is either OK or Will download. The import will fail if there are unmatched references.
For references located on a CLC Genomics Server, the table is empty. The importer can be launched, regardless of whether the correct references are selected, but it leads to an error in cases where they are not.
Output options
In the 'Result handling' wizard step, the downloaded reference sequences can be saved using the option Save downloaded reference sequences if the option Download references when link available was selected in the 'Set parameters' wizard step.
One sequence list per read group is created. Mapping information in the CRAM file is disregarded.
