Fasta read files
The Fasta Read Files importer is designed to import large volumes of read data such as high-throughput sequencing data (NGS reads) in fasta format. Uncompressed files as well as files compressed using gzip (.gz), zip (.zip) or bzip2 (.bz2) can be provided as input. When using the importer, read names can be included but descriptions from the fasta files are ignored.
To launch the Fasta Read Files importer, go to:
Import (![]() ) | Fasta Read Files (
) | Fasta Read Files (![]() ).
).
This opens a dialog where files can be selected and import options specified (figure 7.21).
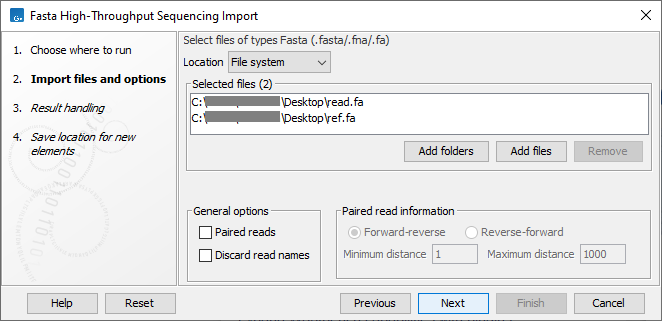
Figure 7.21: Importing data in fasta format.
For import of other fasta format data, such as reference sequences, please use the Standard Import (![]() ) instead (see Standard import), as this import format also includes the descriptions. To have a reference in track format, please use Tracks (
) instead (see Standard import), as this import format also includes the descriptions. To have a reference in track format, please use Tracks (![]() ) and set the "Type of files to import" to FASTA.
) and set the "Type of files to import" to FASTA.
This data type can also be imported using the on-the-fly import functionality available in workflows, described in Launching workflows individually and in batches.
The General options to the left are:
- Paired reads. For paired import, the CLC Genomics Workbench expects the forward reads to be in one file and the reverse reads in another. The CLC Genomics Workbench sorts the files before import and then assumes that the first and second file belong together, and that the third and fourth file belong together etc. At the bottom of the dialog, you can choose whether the ordering of the files is Forward-reverse or Reverse-forward. As an example, you could have a data set with two files:
sample1_fwdcontaining all the forward reads andsample1_revcontaining all the reverse reads. In each file, the reads have to match each other, so that the first read in thefwdlist should be paired with the first read in therevlist. Note that you can specify the insert sizes when importing paired read data. If you have data sets with different insert sizes, you should import each data set individually in order to be able to specify different insert sizes. Read more about handling paired data. - Discard read names. Selecting this option saves disk space. Names of individual sequences are often irrelevant in large datasets.
Click Next to adjust how to handle the results. We recommend choosing Save in order to save the results directly to a folder, since you probably want to save anyway before proceeding with your analysis. There is an option to put the import data into a separate folder. This can be useful for better organizing subsequent analysis results and for batch processing.
