Running Demultiplex Reads in workflows
A number of sequencing kits offer indexing kits where barcodes are coded into plate wells, e.g. the QIAseq UPX sequencing kits. Often each individual well has a specific barcode that is always placed in the plate matrix at the same position. Usually different plates are available and plexing can be done for up to 386 wells (4 x 96 different plate well designs). When running these protocols demultiplexing directly on the sequencing machine is not appropriate and hence the demultiplexing step needs to be done using the Demultiplex Reads tool instead.
When analyzing multiple samples it is not always that the full 96 samples plate is used. This result in empty wells that needs to be removed. When running the Demultiplex Reads tool in a workflow you get the option to load or import your barcode table, have a preview of what wells have been used and you are given the option to remove the barcodes (wells in the plate) that have not been used, or that for some reason did not produce any or only few reads.
An example of a plate design as well as the barcode preview is shown in figure 28.26 and figure 28.27. A Delete button is present and will remove highlight barcodes.
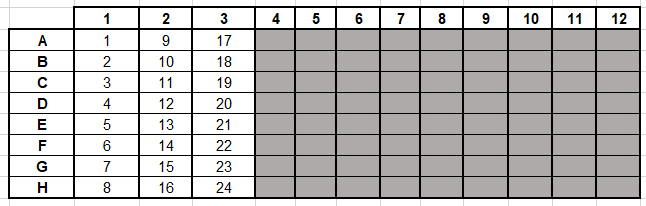
Figure 28.26: The sample design organisation of an experiment not using all the wells in the plate.
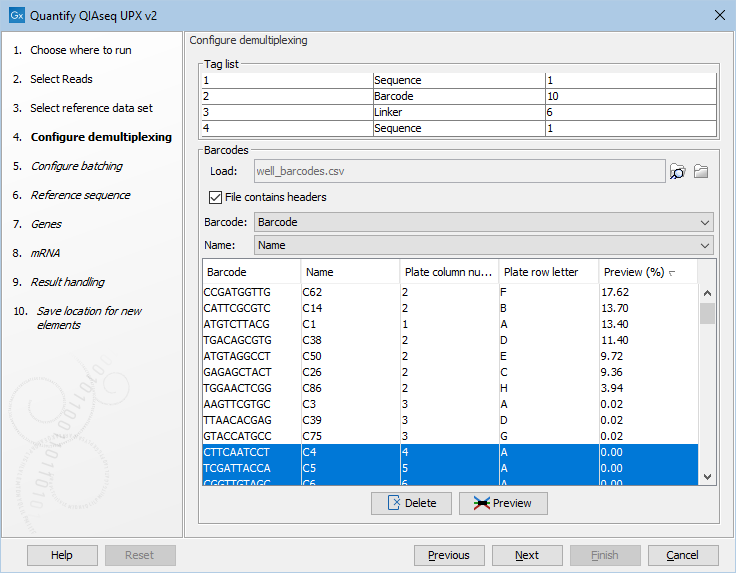
Figure 28.27: A preview of the barcode table that was loaded. Barcodes can be deleted by selecting them, as shown here, and clicking on the Delete button.
Use the preview functionality to identify barcodes matching reads. When the Preview column is empty you can fill in the percentages by selecting the preview button and the selecting the reads. Use sorting by ticking the Preview column header to rank the barcodes according to percent reads per barcode. It will now be easy to select wells that are not used or have low quality.
You can also just deselect any barcode/well that was not used. For this sorting on plate column number and plate row number as illustrated in figure 28.27 could be helpful.
Note: When reads output from Demultiplex Reads in a workflow should be further processed, an Iterate element must be connected to the 'Demultiplexed Reads' output channel. The next downstream analysis element should be connected to the output channel of that Iterate element. Each set of reads is then processed individually, either until the workflow ends or until a Collect and Distribute element is encountered.
