Import Expression Data
Import Expression Data imports expression data from a tabular file. The importer produces one expression track per sample.
To run the importer, go to:
Tools | RNA-Seq and Small RNA Analysis (![]() ) | RNA-Seq Tools (
) | RNA-Seq Tools (![]() ) | Import Expression Data (
) | Import Expression Data (![]() )
)
The following options can be configured (figure 33.66):
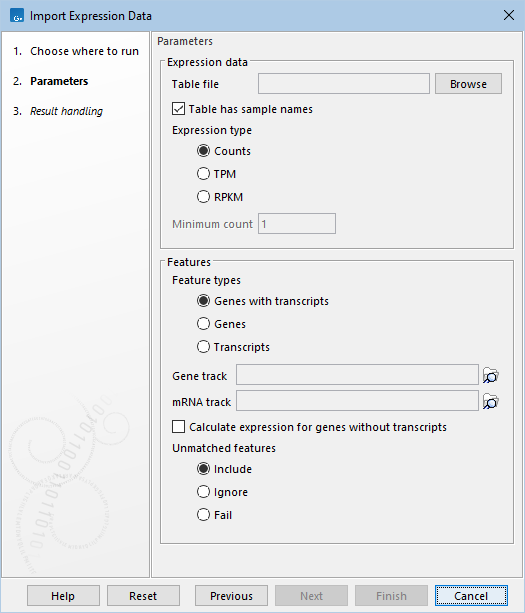
Figure 33.66: Configurable options for "Import Expression Data".
- Table file An Excel, CSV or TSV file containing the expression data (figure 33.67) where:
- Columns represent samples and rows represent genes.
- Feature (gene or transcript) names/IDs are in the first column.
- Feature IDs represent a unique type of identifier, such as Ensembl or geneID.
- Expression values are non-negative and of the same type, such as raw counts or TPM.
- Table has sample names When checked, sample names are read from the first row in the file and are used as the names for the output expression tracks. Otherwise, the expression tracks are named based on the table file name.
- Expression type The type of expression values contained in the file:
- Counts The raw counts. This is recommended when available.
- TPM Transcripts per million.
- RPKM Reads Per kilobase of exon model per million mapped reads.
- Minimum count The smallest raw count found in the original expression data, typically 1 for unfiltered data. It is used for calculating the counts in the expression tracks when the expression type is TPM or RPKM.
If the minimum count is unknown, it should ideally be set to be in the same order of magnitude as the original raw counts. Setting the minimum count too low can lead to a loss of precision, while setting it too high can create a false sense of precision, causing genes with equal expression levels to appear different due to small variations in exon lengths.
- Feature types The types of features for which expression values are contained in the file:
- Genes with transcripts The feature names/IDs are matched against the gene track. The corresponding transcripts are used to calculate exon lengths when converting between counts and TPM/RPKM. The importer outputs gene expression tracks.
- Genes The feature names/IDs are matched against the gene track. The gene lengths are used when converting between counts and TPM/RPKM. The importer outputs gene expression tracks.
- Transcripts The feature names/IDs are matched against the mRNA track. The exon lengths are used when converting between counts and TPM/RPKM. The importer outputs transcript expression tracks.
- Gene track The gene track used for matching the genes names/IDs in the file.
- mRNA track The mRNA track used for converting between counts and TPM/RPKM or for matching the transcript names/IDs in the file.
- Calculate expression for genes without transcripts When checked, the gene lengths are used when converting between counts and TPM/RPKM for genes without a corresponding transcript. This option is only applicable when the feature type is set to Genes with transcripts.
- Unmatched features
When matching feature names/IDs from the file against the gene or mRNA track, some features may remain unmatched either because they are not found in the track or because they are ambiguous, matching multiple features in the track. The following options are available:
- Include Unmatched features are included without information on chromosome or genomic position (region). This option can only be used when importing raw counts.
- Ignore Unmatched features are ignored.
- Fail Unmatched features cause the importer to fail.
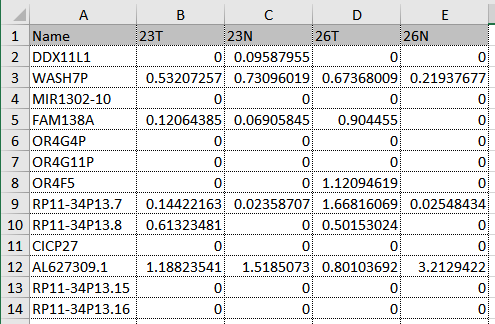
Figure 33.67: Expression data for four samples. The first column contains Ensembl gene names and the expressions are RPKM values.
Importing sample metadata
Certain types of analysis, such as Differential Expression for RNA-Seq, require sample metadata. The sample metadata can be imported into a CLC Metadata Table from an Excel, CSV or TSV file, see Importing metadata for details. Once the metadata is imported, it can be associated with the imported expression tracks, see Associating data elements with metadata for details.
When used in a workflow, Import Expression Data can both import the metadata and associate the resulting CLC Metadata Table with the expression tracks. The CLC Metadata Table is used by downstream tools that require metadata, such as Differential Expression for RNA-Seq. The following additional options can be configured:
- Import metadata Enables import of sample metadata.
- Metadata file An Excel, CSV or TSV file containing the metadata (figure 33.68).
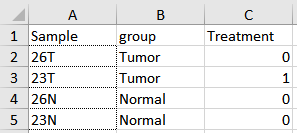
Figure 33.68: Sample metadata for the expression data from figure 33.67.
