Polish Contigs with Reads output
In addition to the sequence list of polished sequences, a summary report is available via the output option Create report.
Polish Contigs with Reads report
An example of the first sections of the Polish Contigs with Reads summary report is shown in figure 35.32.
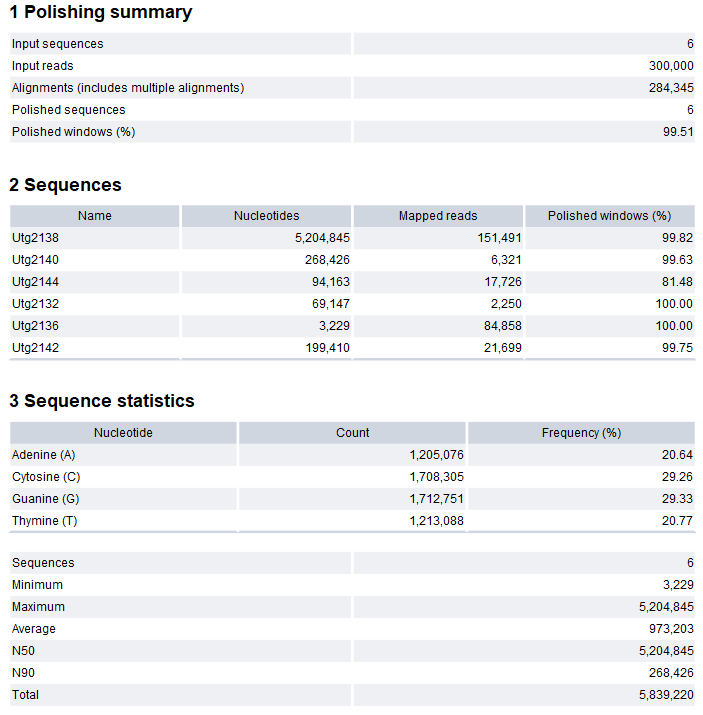
Figure 35.32: Polish Contigs with Reads report
The report contains the following information:
- Polishing summary
- Input sequences. Number of target input sequences.
- Input reads. Number of high-quality input reads.
- Alignments (includes multiple alignments). The number of alignments between target sequences and high-quality reads.
- Polished sequences. Number of sequences that were polished.
- Polished windows (%). The percentage of windows that were polished.
- Sequences. For each target input sequence, the following information is given:
- Name. Name of the sequence.
- Nucleotides. Number of nucleotides in the sequence.
- Mapped reads. Number of high-quality reads that mapped to the sequence.
- Polished windows (%). The percentage of windows that were polished.
- Sequence statistics
- Sequences. Number of sequences.
- Minimum, Maximum, Average. Minimum, maximum and average sequence length.
- N50. The length of the shortest sequence in sets of sequences of equal length or longer, where the summed length of sequences is at least 50% of the total sequence length.
- N90. The length of the shortest sequence in a set of sequences of equal length or longer, where the summed length of sequence is at least 90% of the total sequence length. N90 will be equal to or smaller than N50.
- Total. The number of bases in the sequences.
- Sequence length distribution. A graph depicting the number of sequences found at a specific length.
