Motif Search
Motif Search takes as input sequences (![]() ) (
) (![]() ) (
) (![]() ), sequence lists (
), sequence lists (![]() ) (
) (![]() ), or alignments (
), or alignments (![]() ), and outputs motif search results containing the detected matches.
), and outputs motif search results containing the detected matches.
To run the tool, go to:
Tools | Classical Sequence Analysis (![]() ) | General Sequence Analysis (
) | General Sequence Analysis (![]() )| Motif (
)| Motif (![]() ) | Motif Search (
) | Motif Search (![]() )
)
The following options can be configured (figure 18.28):
- Search using Choose whether to search for a single motif by selecting one of the available motif types, or to search using a Motif list.
- Motif The motif sequence to search for, if searching for a single motif. For syntax details, see Motif types.
- Motif list The motif lists (
 ) to use when searching with a list.
) to use when searching with a list.
- Accuracy (%) The percentage of residues that must match the motif. This option only impacts simple motifs. Indels are not supported.
Ambiguous characters are considered different if they do not have any residues in common. For example, N matches any nucleotide and R matches nucleotides A,G, while X matches any amino acid and Z matches amino acids E,Q.
- Include negative strand When checked, the reverse complement sequence of the motifs is also searched on the negative strand. This option is only available for nucleotide sequences.
- Exclude ambiguous region When checked, motifs are not searched for in N-regions. Additionally, when a motif matches to an N, it is treated as a mismatch.
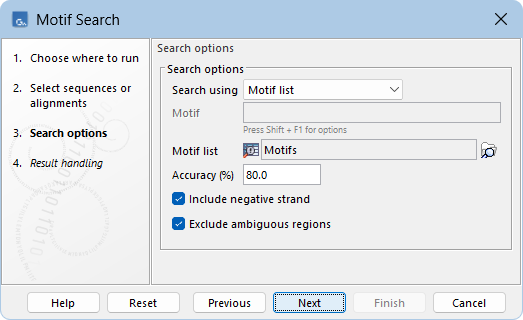
Figure 18.28: Search for several motifs using a motif list.
In the "Result handling" wizard, the following options can be configured:
- Create report When checked, a report is produced summarizing the detected motif matches (figure 18.29).
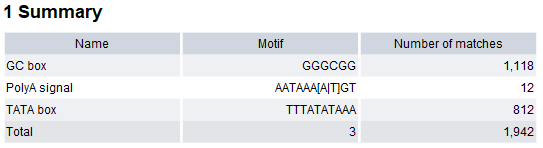
Figure 18.29: Report produced by Motif Search. - Create table When checked, a table is produced containing details of all detected motif matches (figure 18.30):
- Sequence The name of the sequence where the match is found.
- Start and End Positions of the match within the sequence.
- Match The sequence segment that matches the motif. For simple motifs with accuracy below 100%, matching residues are in uppercase, and non-matching residues in lowercase.
- Match length The number of residues in the match.
- Accuracy (%) The percentage of matching residues between the motif and the matched sequence segment.
- Strand Strand where the match occurs.
- Name, Motif, Description, and Annotation type Details from the matching motif.
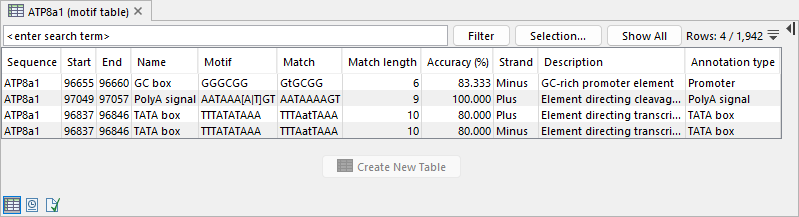
Figure 18.30: Table produced by Motif Search. - Add annotations When checked, annotations are added to the sequences in the input (figure 18.31). The annotation type is determined by the motif, and all motif information is transferred to the annotation. See Working with annotations for more information.
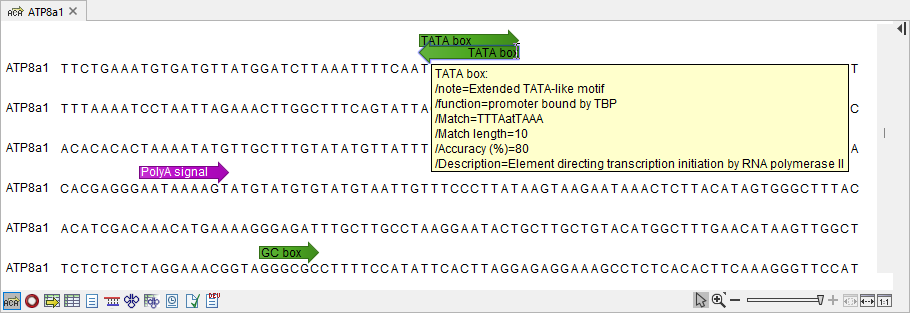
Figure 18.31: Annotations added by Motif Search.
