Prepare Raw Data
The Prepare Raw Data workflow performs quality control and trimming of the sequencing reads.
Launching the workflow
The Prepare Raw Data workflow is available from the Toolbox at:
Workflows | Template Workflows | Preparing Raw Data (![]() ) | Prepare Raw Data (
) | Prepare Raw Data (![]() )
)
The Prepare Raw Data workflow contains two tools, QC for Sequencing Reads and Trim Reads.
Launch the workflow and step through the wizard.
- Select the reads to be processed.
If more than one sequence list has been selected, check Batch to analyze each input separately. This will generate a trimmed sequence list and QC report for each sequence list provided. See Running workflows in batch mode for details.
If multiple sequence lists are input and the workflow is not run in Batch mode, a single sequence list and a single QC report are generated, based on all the reads from all the sequence lists.
- In the next step, choose "Use organization of input data" to specify how to define the batch units.
- Next, you can review the batch units resulting from your selections above.
- In the next step, settings for the Trim Reads tool can be inspected and adjusted (figure 14.97).
If analyzing RNA-Seq data, we recommend trimming polyA tails by checking Trim homopolymers from 3', polyA, and polyT.
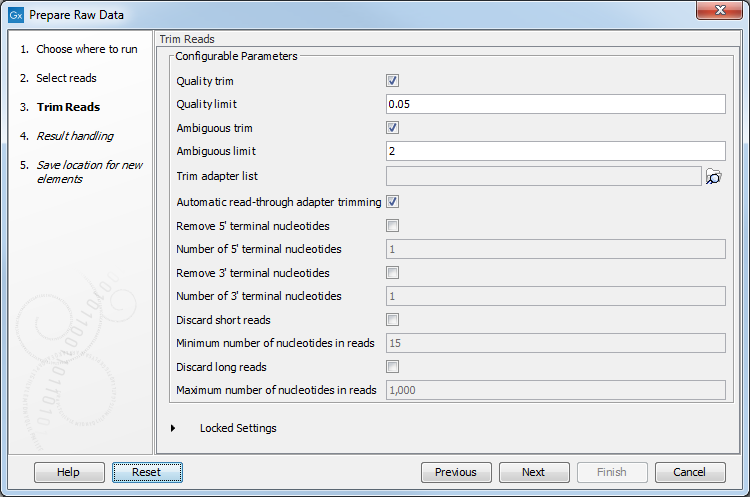
Figure 14.97: Trim Reads settings can be adjusted. - In the next step, you can click on Preview All Parameters to review your settings and specify how results should be handled.
- In the final step, you can choose a location to save the results to.
Generated outputs
The outputs provided by this workflow are:
- Graphical report. See QC for Sequencing Reads for details.
- Supplementary report. See QC for Sequencing Reads for details.
- Trimmed sequences. Sequence list containing trimmed paired reads. See Trim output for details.
- Trimmed (broken pairs). Sequence list containing trimmed broken paired reads. See Trim output for details.
- Trim report. See Trim output for details.
The reports generated should be inspected to determine whether the quality of the sequencing reads and the trimming is acceptable. If the quality is acceptable, the trimmed reads can be used in downstream analyses.
