Download Genomes
Download Genomes provides access to genomes and associated reference data, such as annotations and known variants, from public repositories11.1.
Download Genomes is accessed by selecting the tab with that name, available at the top of the Reference Data Manager.
To open the Reference Data Manager, click on the Manage Reference Data (![]() ) button in the top Toolbar or go to the Utilities menu and select Manage Reference Data (
) button in the top Toolbar or go to the Utilities menu and select Manage Reference Data (![]() ).
).
Under the Download Genomes tab is a list of organisms for which data can be downloaded (figure 11.2). The number in a circle to the left of each organism name indicates how many previously downloaded data sets are available in the CLC_References location for that organism.
After selecting an organism in the list, information about the data available for download is shown in the middle section of the right hand panel, including a name describing the data resource type, a version (if available), the data provider, and the file size.
Note: Most data is supplied as a compressed text file. After download, each file is decompressed and the data is imported. CLC data is compressed by default, but the size of the compressed data after import will generally be different to the size reported for the original data file.
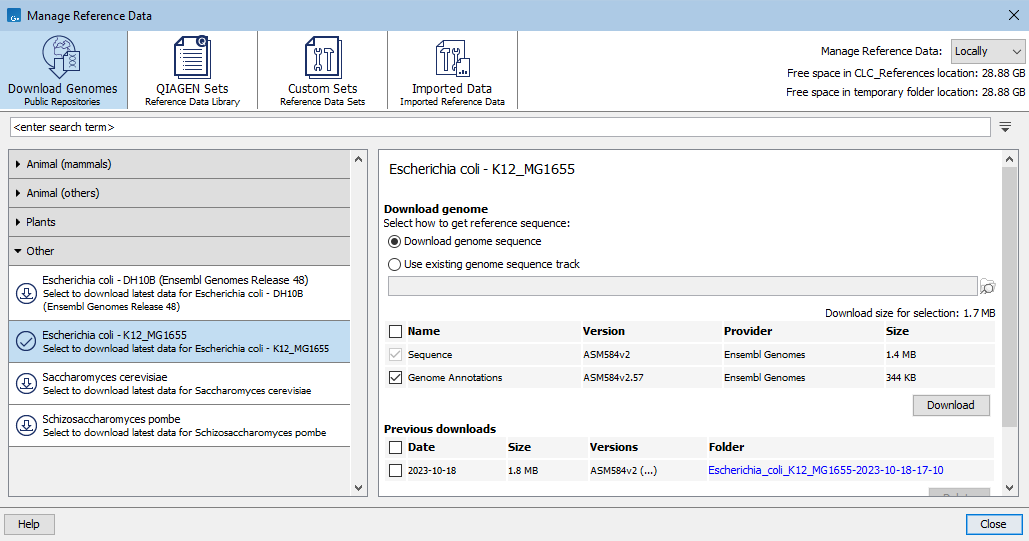
Figure 11.2: Under the Download Genomes tab, reference sequences and associated reference data for a variety of organisms can be downloaded from public repositories.
Data selected will be imported as tracks. When importing annotations, variants, and other types of data, a genome sequence track is needed to provide the genomic coordinates that all the imported tracks will share. The radio buttons under "Select how to get reference sequence:" offer the choice of using the track that will be created from sequence data being downloaded ("Download genome sequence") or using a sequence track already available ("Use existing genome sequence track").
Download and import
Select the data types of interest and click the Download button. The data will then be downloaded from the public repository and subsequently imported.
When reference data is stored on a CLC Server with grid nodes, the grid preset to use to download data can be specified via a drop-down menu to the left of the Download button (figure 11.3).
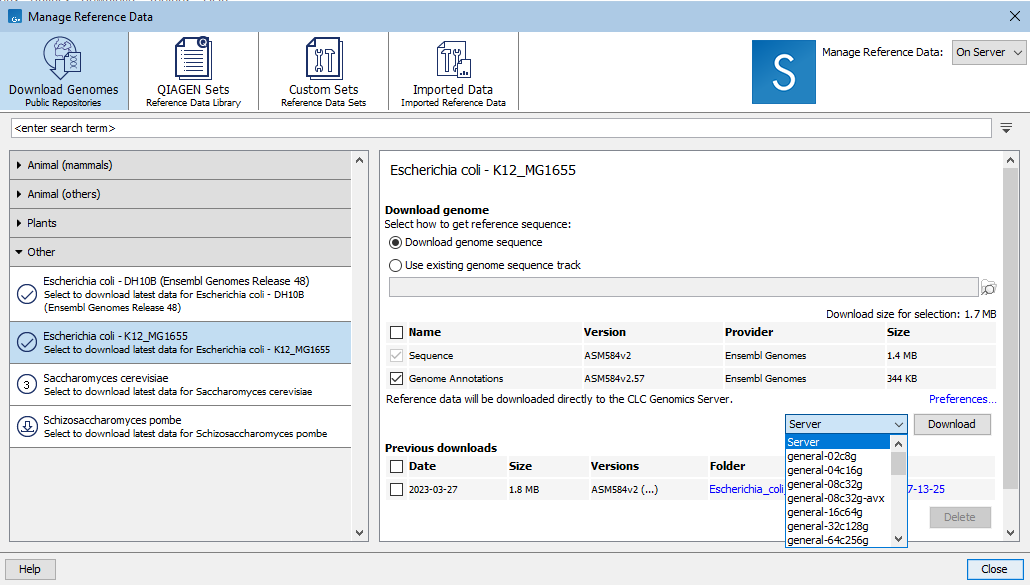
Figure 11.3: When the "On Server" option, top right, is selected and grid presets are configured on the CLC Server, a grid preset to use for the download can be selected.
When genome annotations are imported, a track is created for each feature type present in the annotation file.
In addition, depending on the annotation source, tracks containing sets of related features may also be created. These tracks have names ending in "(RNA)" and "(Gene)" (figure 11.4) and can be useful for some types of analyses, e.g. RNA-Seq. The (RNA) track contains entries for all "RNA" type annotations. I.e. all the children of "mature_transcript", which is the parent of "mRNA", which is the parent of the "NSD_transcript". The (Gene) track contains genes and gene-like annotation types, such as ncRNA_gene, plastid_gene, and tRNA_gene.
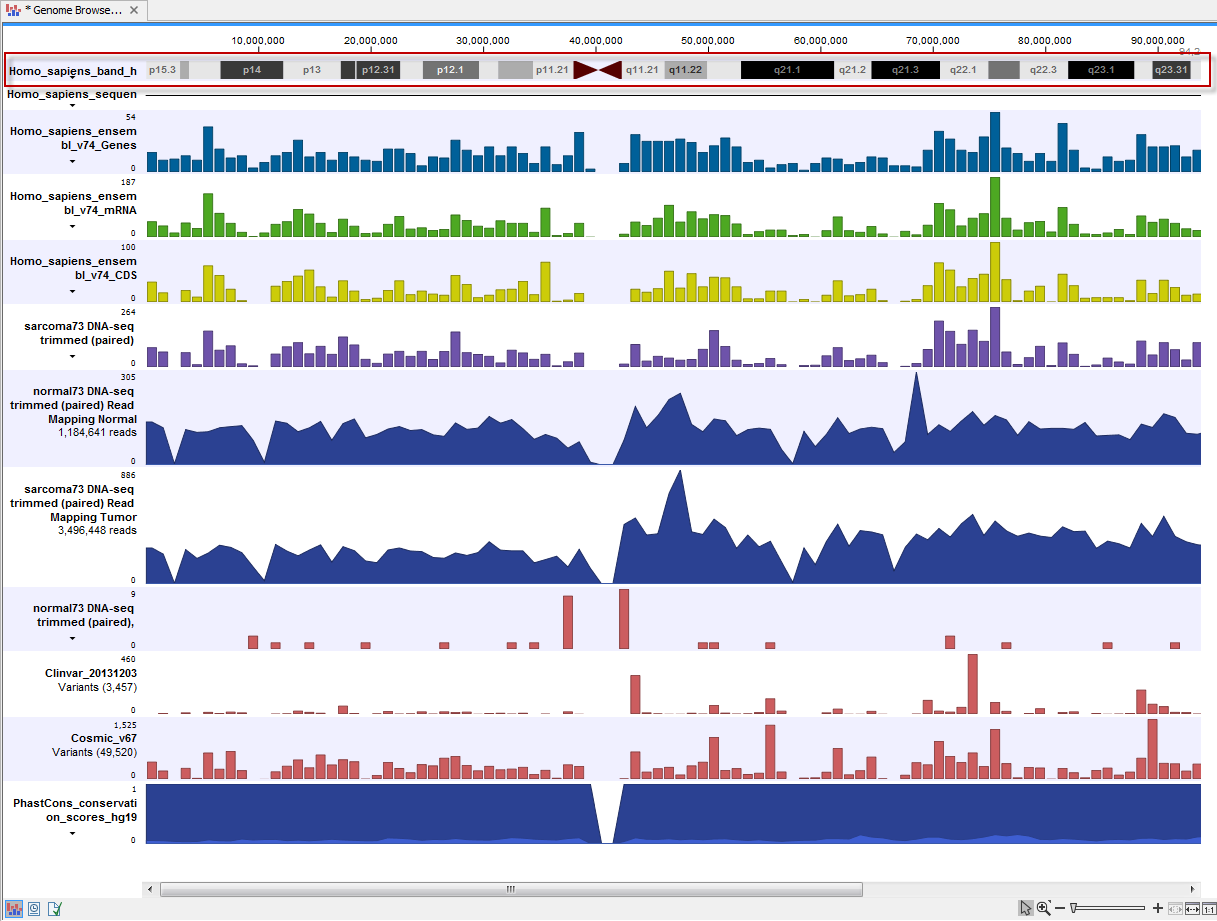
Figure 11.4: A track list containing some of the tracks created by Download Genomes for the Homo Sapiens hg38 genome.
Finding and deleting data imported using Download Genomes
The imported tracks are stored under the CLC_References location, under the "Genomes" folder. Each downloaded set is placed in a folder with a name containing the species name and the date of the download.
To easily navigate to the imported tracks in the Navigation Area, click on a link under the Folder column in the "Previous downloads" section at the bottom of the right hand panel in the Reference Data Manager. This moves the focus in the Navigation Area to the location of the downloaded data.
Hovering the mouse cursor over information in the Versions column in the "Previous downloads" section reveals a tooltip with a list of the resources downloaded and, if available, the version of each (figure 11.2).
To delete data downloaded using Download Genomes, select one or more entries in the list of previous downloads and click on the Delete button. When reference data is stored on a CLC Server, you need be logged in from the Workbench as an administrative user to delete reference data.
Searching for data available under the Download Genomes tab
The search field under the top toolbar can be used to search for terms in the names of organisms, resources, and resource providers. To search for just an exact term, put the term in quotes.
The results include the name of the element or set the term was found in, followed by the tab it is listed under, in parentheses. Hover the cursor over a hit to see what aspect of the result matched the search term (figure 11.5). Double-click on a search result to open it.
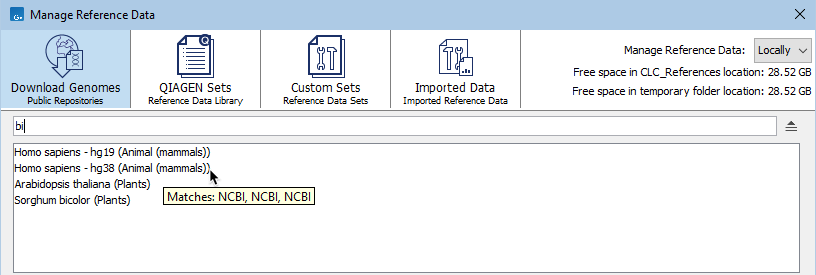
Figure 11.5: When the Download Genomes tab is selected, terms entered in the search field are searched for in the names of organisms, resources, and resource providers. Hovering the cursor over a hit reveals a tooltip with information about the match.
Footnotes
- ... repositories11.1
- The data listed under the Download Genomes tab is not provided by nor hosted by QIAGEN.
