Normalization and clustering
The expression values are filtered and normalized as follows:
- Features with zero expression across all samples, or invalid values (NaN or +/- Infinity), are removed.
- logCPM values are calculated for each feature.
- A Z-Score normalization is applied on the logCPM values.
The samples and features, as relevant, are hierarchically clustered based on the similarity of their expression profiles, as follows:
- Create clusters containing one sample/feature.
- Calculate the distances between all clusters.
- Merge the two closest clusters into one.
- Repeat until only one cluster remains, containing all the samples/features.
The hierarchical cluster forms tree structures displayed along the rows and columns of the heat map. The tree branch lengths represent the distances between clusters.
The distance between two clusters is determined using one of the following linkage types:
- Single linkage. The distance between the two closest samples/features in the two clusters.
- Average linkage. The average distance between samples/features in the first cluster and samples/features in the second cluster.
- Complete linkage. The distance between the two farthest samples/features in the two clusters.
The distance between two samples/features is calculated using one of the following distance measures:
- Euclidean distance. The length of the segment connecting two points. If
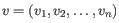 and
and
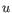 , then the Euclidean distance between
, then the Euclidean distance between 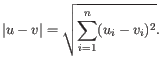 and
and 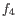 is
is
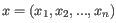
- Manhattan distance. The distance between two points measured along axes at right angles. If
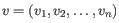 and
and
 , then the Manhattan distance between
, then the Manhattan distance between 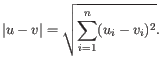 and
and 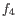 is
is
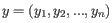
- 1 - Pearson correlation. The Pearson correlation coefficient between
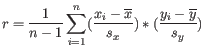 and
and
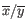 is defined as
where
is defined as
where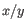
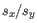 and
and 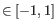 are the average and sample standard deviation, respectively, of the values in
are the average and sample standard deviation, respectively, of the values in 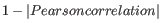 values.
values.
The Pearson correlation coefficient ranges from -1 to 1, with high absolute values indicating strong correlation, and values near 0 suggesting little to no relationship between the elements.
Using 1 - | Pearson correlation | as the distance measure ensures that highly correlated elements have a shorter distance, while elements with low correlation are farther apart.
