- Defining batch units for workflows with Iterate elements
- Importing sequence data with sample information
- Renaming Iterate elements
- Further configuring Iterate elements
- Configuring Collect and Distribute elements
Iterate and Collect and Distribute elements
Iterate (![]() ) and Collect and Distribute (
) and Collect and Distribute (![]() ) elements are used to control how data is grouped for analysis.
) elements are used to control how data is grouped for analysis.
- Iterate elements are placed at the top of a branch of a workflow that should be run multiple times, using different inputs in each run. The sets of data to use in each run are referred to as "batch units" or, sometimes, "iteration units".
- Collect and Distribute elements are, optionally, placed downstream of an Iterate element, where they collect outputs from the upstream iteration block (see below) and distribute them as inputs to downstream analyses.
The RNA-Seq and Differential Gene Expression Analysis template workflow, distributed with the CLC Genomics Workbench (https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=RNA_Seq_Differential_Gene_Expression_Analysis_workflow.html) is an example of a workflow that includes each of these control flow elements.
The steps between an Iterate element and a Collect and Distribute element are referred to as an "iteration block". The workflow in figure 14.48 contains a single iteration block (shaded in turquoise), where steps within that block are run once per batch unit. The Collect and Distribute element (renamed to Collect Expressions) collects all the results from the iteration block and sends it as input to the next stage of the analysis (shaded in purple).
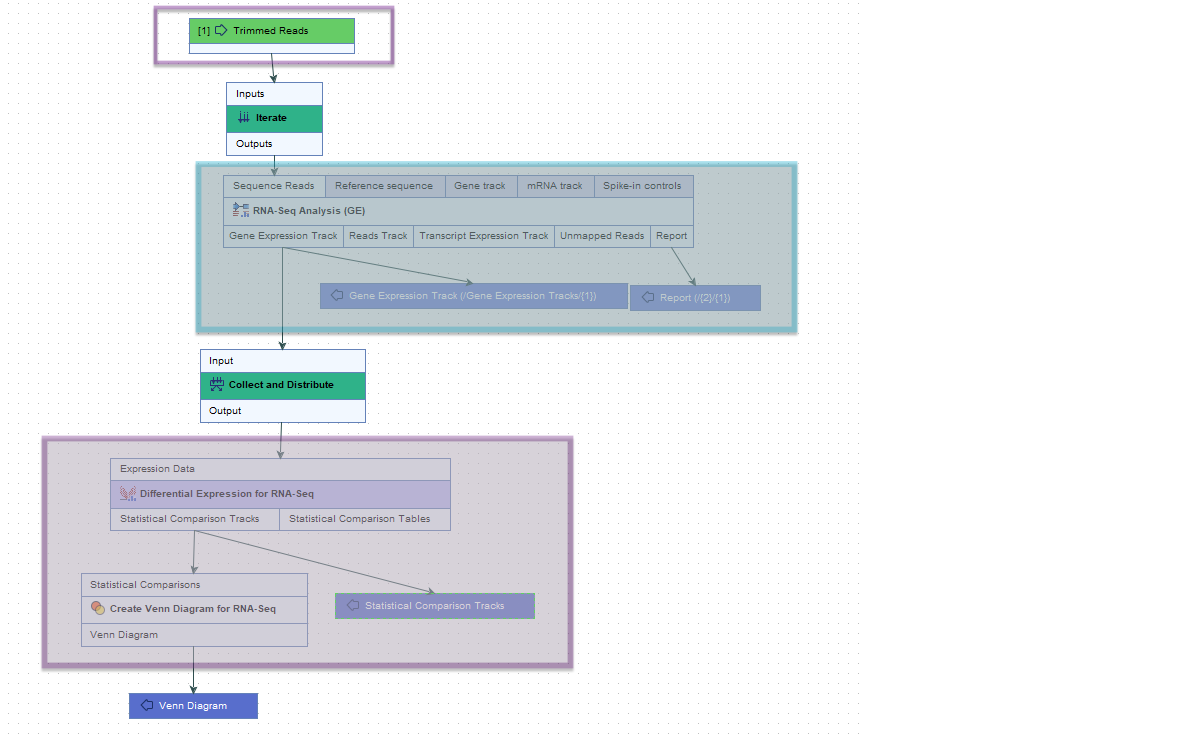
Figure 14.48: The roles of the Iterate and Collect and Distribute control flow elements are highlighted in the context of RNA-Seq and differential expression analyses.
RNA-Seq Analysis lies downstream of an Iterate element, within an iteration block (shaded in turquoise). It will thus be run once per batch unit.
Differential Expression for RNA-Seq lies immediately downstream of a Collect and Distribute element (renamed to Collect Expressions), and is sent all the expression results from the iteration block as input for a single analysis.
Defining batch units for workflows with Iterate elements
Workflow elements downstream of an Iterate element are run once for each batch unit. Details about defining batch units when launching workflows is described at Running workflows in batch mode.
Running a workflow with a single Iterate element at the top of a workflow, no downstream Collect and Distribute element, and a single Input element is equivalent to running a similar workflow without the Iterate element in Batch mode. Setting up batch units in this situation is described in Batch processing.
Importing sequence data with sample information
A workflow containing just an Input, Output and Iterate element can be a useful tool to create a CLC Metadata Table with sample information and data elements associated with the relevant rows. This can then be used when launching tools and workflows requiring metadata. A template workflow with this design, Import with Metadata, is provided in the Preparing Raw Data template workflow folder, available under the Workflows menu, and is described in Import with Metadata.
Renaming Iterate elements
Providing meaningful names to Iterate elements can help at both the workflow design stage and also when launching the workflow.
The Rename option is available in the menu that appears when you right-click on a workflow element.
Iterate element names are included in the workflow launch wizard in the following steps:
- Configure batching: The name of Iterate elements are provided in association with the drop-down list of column names in the metadata provided. A meaningful Iterate element name can thus help guide the choice of relevant metadata to group the inputs into batch units (figure 14.49).
- Batch overview: There is a column for each Iterate element (figure 14.50). Meaningful names can thus make it easier to review batch unit organization critically when launching the workflow.
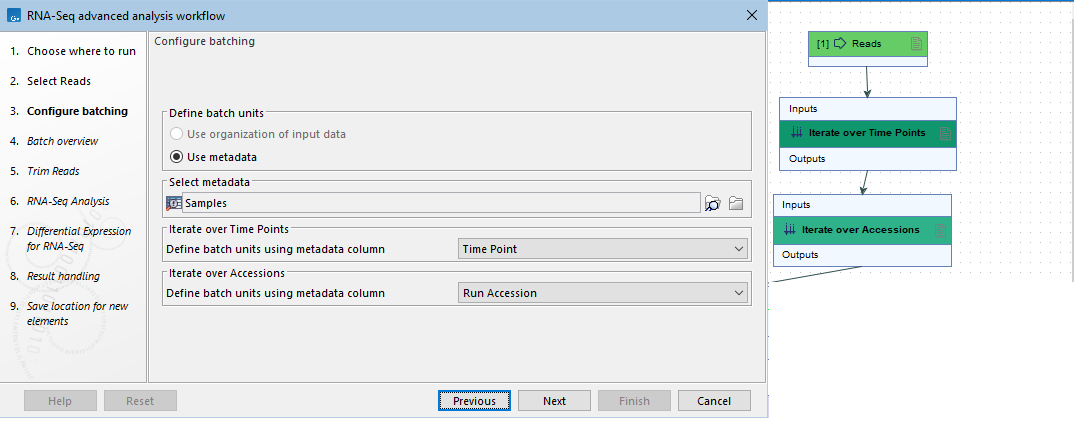
Figure 14.49: The two Iterate elements in this workflow (right) have been renamed. Their names are included in the "Configure batching" wizard step in the launch wizard (left).
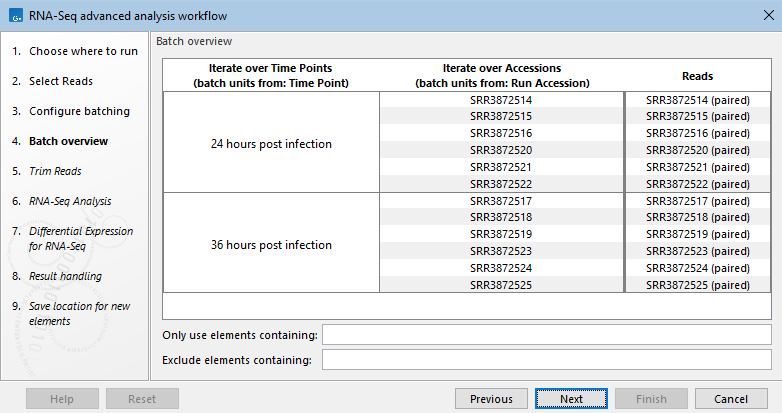
Figure 14.50: The batch overview for a workflow with two Iterate elements. The names assigned to the two columns containing the batch unit organization are the names of the corresponding Iterate elements.
Further configuring Iterate elements
Double-clicking on an Iterate element opens the configuration dialog, which contains the options listed below (figure 14.51). The default settings are relevant for most uses of the Iterate element.
- Number of coupled inputs The number of separate inputs for each given iteration. These inputs are "coupled" in the sense that, for a given iteration, particular inputs are used together. For example, when sets of sample reads should be mapped in the same way, but each set should be mapped to a particular reference (figure 14.52).
- Error handling Specify what should happen if an error is encountered. The default is that the workflow should stop on any error. The alternative is to continue running the workflow if possible, potentially allowing later batches to be analyzed even if an earlier one fails.
- Metadata table columns If the workflow is always run with metadata tables that have the same column structure, then it can be useful to set the value of the column titles here, so the workflow wizard will preselect them. The column titles must be specified in the same order as shown in the workflow wizard when running the workflow. Locking this parameter to a fixed value (i.e. not blank) will require the definition of batch units to be based on metadata. Locking this parameter to a blank value requires the definition of batch units to be based on the organization of input data (and not metadata).
- Primary input If the number of coupled inputs is two or more, then the primary input (used to define the batch units) can be configured using this parameter.
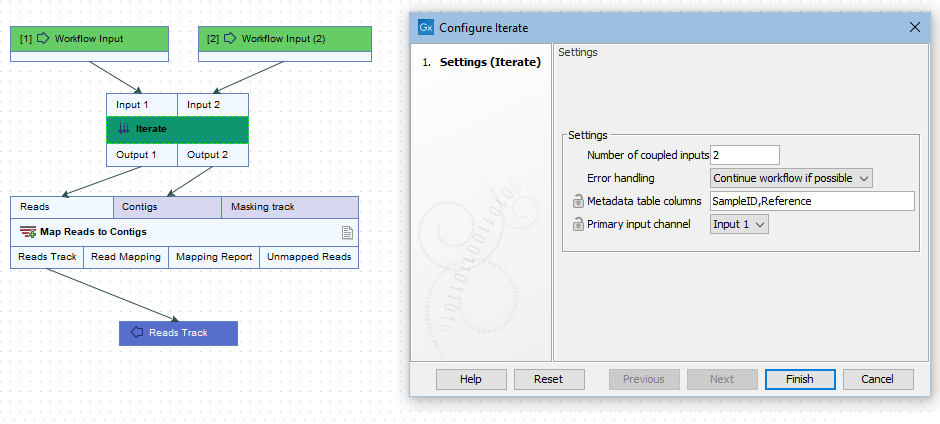
Figure 14.51: The number of coupled inputs in this simple example is 2, allowing each set of sample reads to be mapped to a paticular reference, rather than using the same reference for all iterations.
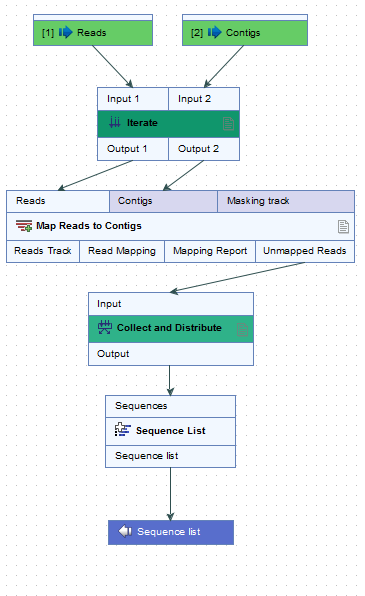
Figure 14.52: Reads can be mapped to specified contigs due to the 2 input channels of the Iterate element. Using this design, a single sequence list containing all the unmapped reads from all the initial inputs is generated. That would not be possible without the inclusion of the Iterate and Collect and Distribute elements.
Configuring Collect and Distribute elements
By default, a Collect and Distribute element has one output channel. In this case, all results from the iteration block are collected and passed to downstream steps of the workflow.
More than one output channel can be configured by entering terms in a comma separated list in the Outputs field (figure 14.53). The number of terms determines the number of output channels. Connections between these output channels and input channels of downstream elements determine how data should be distributed in the following stage of the workflow.
If the Collect and Distribute element has more than one output channel, the path taken by a given element is determined by the value in the metadata column specified when launching the workflow. This column can be preconfigured in the Group by metadata column setting.
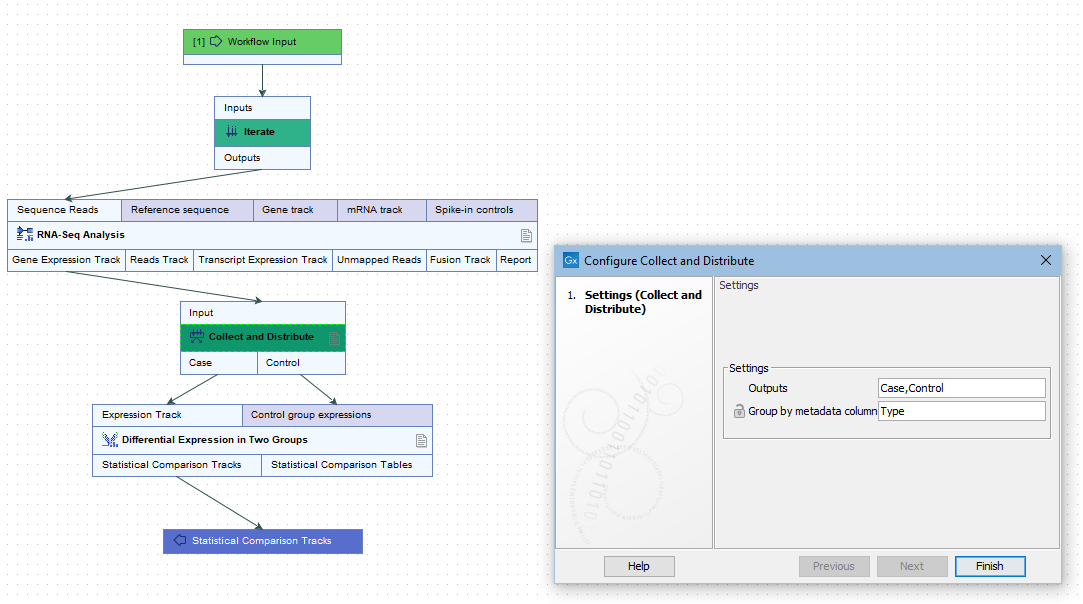
Figure 14.53: A comma separated list of terms in the Outputs field of the Collect and Distribute element defines the number of output channels and their names.
For example, when launching the workflow in figure 14.54, a metadata column called "Type" was specified for defining which samples were cases and which were controls. The iteration units were defined by the contents of the "ID" column (figure 14.55).
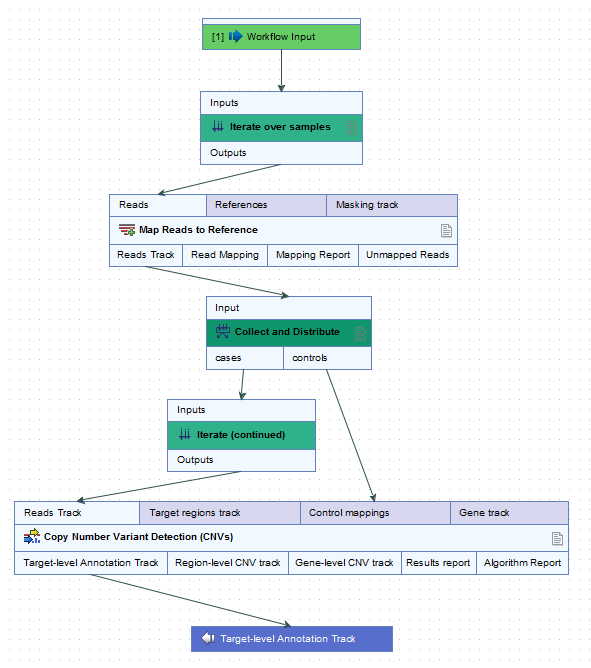
Figure 14.54: In this workflow, each case sample is analyzed against all of the control samples.
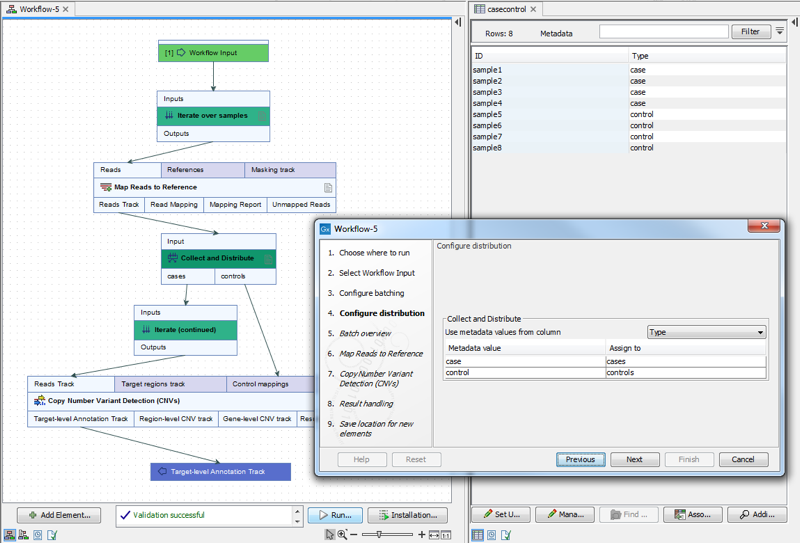
Figure 14.55: Contents of the metadata column "Type" define which samples are cases and which are controls. Iteration units are defined by the contents of the "ID" column.
