Special notes upgrading to Genomics Workbench 7.5
This section is a special note on upgrading to CLC Genomics Workbench 7.5 and CLC Genomics Server 6.5. This is intended for those upgrading from earlier versions and will provide information about how this change affects both existing and new data.
With the new version we have introduced three new variant callers: "Basic Variant Detection", "Fixed Ploidy Variant Detection", and "Low Frequency Variant Detection" (see Variant Detectors - overview). The Basic Variant Detection and Fixed Ploidy Variant Detection are similar in nature to the Quality-based and Probabilistic Variant Detection tools respectively. The main difference is that all filters, previously employed in either the Quality-based or Probabilistic Variant Detector, are now available in all three variant callers, in addition to a new filter: the relative read direction filter. The changes relating to the filters have some few consequences, and care must be taken when attempting to run the new variant callers with parameters that are equivalent to those used in the old ones (see figure 27.49) for details about the Fixed Ploidy Variant Detection and figure 27.48 for details about the Basic Variant Detection).
In particular:
- Regions with coverage above a certain threshold can be masked out in the new callers. To avoid this, you should set the 'Maximum coverage' parameter as high as the maximum coverage observed in the read mapping.
- A parameter (maximum number of gaps and mismatches) has been deleted from the quality filter. This parameter made the algorithm ignore reads with more than the specified number of nucleotide differences to the reference, and had the unfortunate effect that read aligned across longer deletions or insertions would not be considered, which in turn meant that these would be missed. The fact that this parameter has been dropped will imply that you cannot expect to get the same variant calls with the basic variant caller as with the quality-based variant caller, unless the value in the Quality-based variant caller has been set to the maximum read length.
- The Fixed Ploidy Variant Detection algorithm has a 'minimum frequency' parameter that will remove variants with a frequency below the specified threshold. In order for that parameter to have no effect you should set it to zero.
- The Fixed Ploidy variant caller by default employs the new 'Relative read direction filter'. This filter is less stringent than the 'read direction filter' which by default is employed by the old Probabilistic variant caller. To obtain similar filtering settings related to read direction, you should either employ none of the read direction filters in the two variant callers, or employ the 'read direction filter' (with the frequency parameter specified to 5% in the Fixed ploidy) in both of them.
- The Basic variant caller by default employs the new 'Relative read direction filter'. This filter is less stringent than the 'read direction filter' which by default is employed by the old Quality-based variant caller. To obtain similar filtering settings related to read direction, you should either employ none of the read direction filters in the two variant callers, or employ the 'read direction filter' (with the frequency parameter specified to 5% in the Basic variant caller) in both of them.
The table shown in figure 27.48 contains a comparison of the Basic Variant Detection tool with the Quality-based Variant Detection tool, whereas the table in figure 27.49 shows a comparison of the Fixed Ploidy Variant Detection tool with the Probabilistic Variant Detection tool.
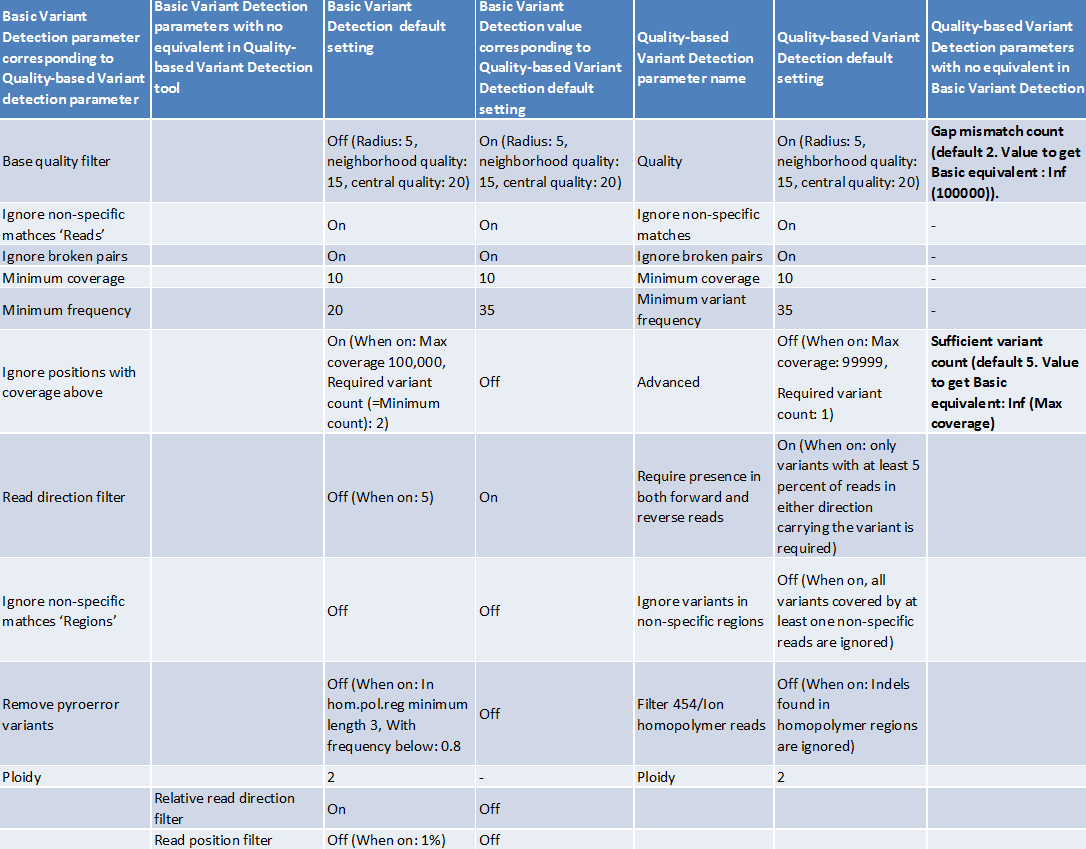
Figure 27.48: Comparison of the Basic Variant Detection tool with the Quality-based Variant Detection tool.
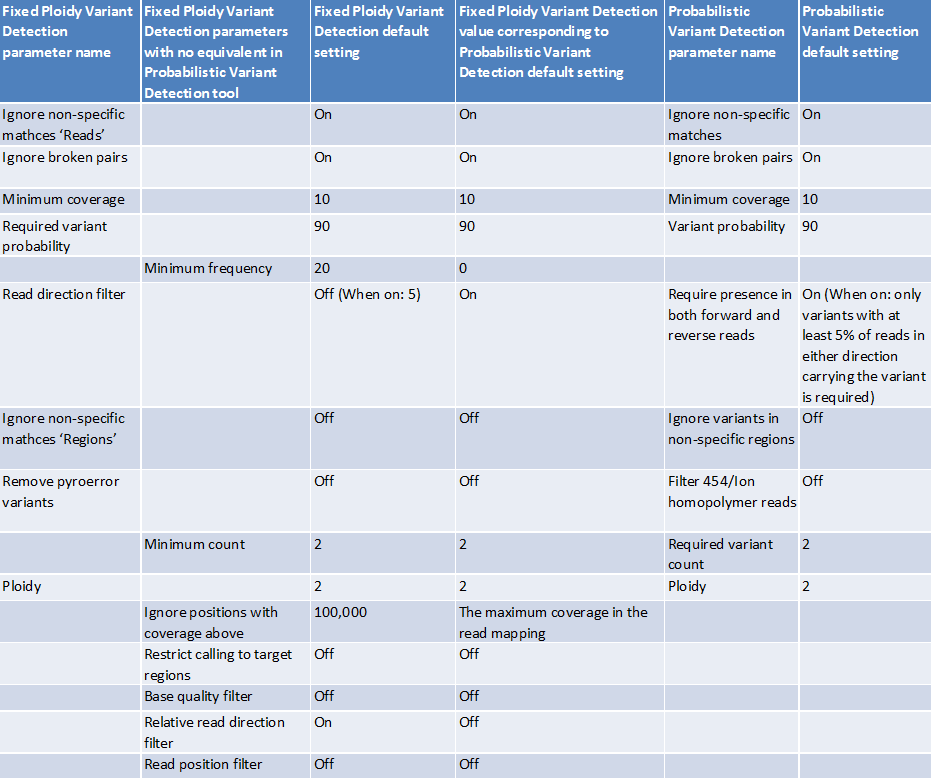
Figure 27.49: Comparison of the Fixed Ploidy Variant Detection tool with the Probabilistic Variant Detection tool.
