Defining reference genome and mapping settings
You are now presented with the dialog shown in figure 27.4.
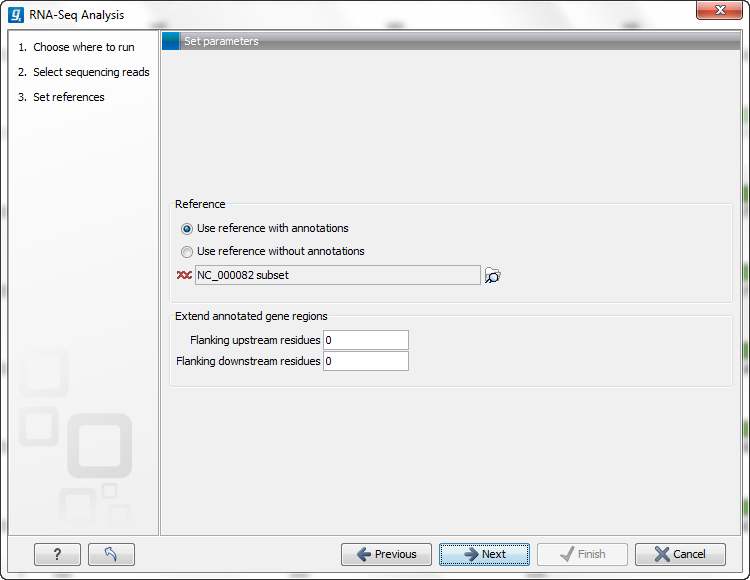
Figure 27.4: Defining a reference genome for RNA-Seq.
At the top, there are two options concerning how the reference sequences are annotated:
- Use reference with annotations. Typically, this option is chosen when you have an annotated genome sequence. Choosing this option means that gene and mRNA annotations on the sequence will be used if you choose the option Eukarotes in the next window. If you choose the option Prokaryotes in the next window, the annotations of type gene only are used. See Finding the right reference sequence for RNA-Seq.
- Use reference without annotations. This option is suitable for situations like mapping back reads to un-annotated EST consensus sequences. The reference in this case is a list of sequences. A common situation is for a multi-fasta file to be imported into the Workbench to be used for this purpose. Each sequence in the list will be treated as a "gene" (or "transcript"). Note that the Workbench uses prokaryote settings here. This means that it does not look for new exons (see Exon discovery) and it assumes that the sequences have no introns).
Just below these two options, you click to select the reference sequences.
Next, you can choose to extend the region around the gene to include more of the genomic sequence by changing the value in Flanking upstream/downstream residues. This also means that you are able to look for new exons before or after the known exons (see Exon discovery).
When the reference has been defined, click Next and you are presented with the dialog shown in figure 27.5.
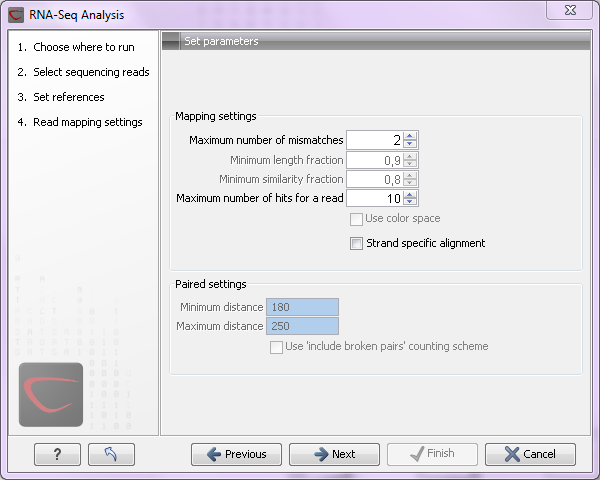
Figure 27.5: Defining mapping parameters for RNA-Seq.
The mapping parameters are:
- Maximum number of mismatches. This parameter is available if you use short reads (shorter than 56 nucleotides, except for color space data which are always treated as long reads). This is the maximum number of mismatches to be allowed. Maximum value is 3, except for color space where it is 2.
- Minimum length fraction. For long reads, you can specify how much of the sequence should be able to map in order to include it. The default is 0.9 which means that at least 90 % of the bases need to align to the reference.
- Minimum similarity fraction. This also applies to long reads and it is used to specify how exact the matching part of the read should be. When using the default setting at 0.8 and the default setting for the length fraction, it means that 90 % of the read should align with 80 % similarity in order to include the read.
- Maximum number of hits for a read.
A read that matches to more distinct places in the references than the 'Maximum number of hits for a read' specified will not be mapped (the notion of distinct places is elaborated below). If a read matches to multiple distinct places, but below the specified maximum number, it will be randomly assigned to one of these places. The random distribution is done proportionally to the number of unique matches that the genes to which it matches have, normalized by the exon length (to ensure that genes with no unique matches have a chance of having multi-matches assigned to them, 1 will be used instead of 0, for their count of unique matches). This means that if there are 10 reads that match two different genes with equal exon length, the two reads will be distributed according to the number of unique matches for these two genes. The gene that has the highest number of unique matches will thus get a greater proportion of the 10 reads.
Places are distinct in the references if they are not identical once they have been transferred back to the gene sequences. To exemplify, consider a gene with 10 transcripts and 11 exons, where all transcripts have exon 1, and each of the 10 transcripts have only one of the exons 2 to 11. Exon 1 will be represented 11 times in the references (once for the gene region and once for each of the 10 transcripts). Reads that match to exon 1 will thus match to 11 of the extracted references. However, when transferring the mappings back to the gene it becomes evident that the 11 match places are not distinct but in fact identical. In this case the read will not be discarded for exceeding the maximum number of hits limit, but will be mapped. In the RNA-seq action this is algorithmically done by allowing the assembler to return matches that hit in the 'maximum number of hits for a read' plus 'the maximum number of transcripts' that the genes have in the specified references. The algorithm post-processes the returned matches to identify the number of distinct matches and only discards a read if this number is above the specified limit. Similarly, when a multi-match read is randomly assigned to one of it's match places, each distinct place is considered only once.
- Strand-specific alignment. When this option is checked, the reads will only be mapped in their forward orientation (genes on the minus strand are reverse complemented before mapping). This is useful in places where genes overlap but are on different strands because it is possible to assign the reads to the right gene. Without the strand-specific protocol, this would not be possible (see [Parkhomchuk et al., 2009]).
Subsections
