SAM and BAM mapping files
The CLC Genomics Workbench supports import and export of files in SAM (Sequence Alignment/Map) and BAM format which are generic formats for storing large nucleotide sequence alignments. Read more and see the format specification at http://samtools.sourceforge.net/.
Please note that the CLC Genomics Workbench also supports SAM and BAM files from Complete Genomics.
For a detailed explanation of the SAM and BAM files exported from CLC Genomics Workbench, please see SAM/BAM export format specification.
The idea behind the importer is that you import the sam/bam file which includes all the reads and then you specify one or more reference sequences which have already been imported into the Workbench. The Workbench will then combine the two to create a mapping result (![]() ) or mapping tables (
) or mapping tables (![]() ). To import a SAM or BAM file:
). To import a SAM or BAM file:
File | Import (![]() ) | SAM/BAM Mapping Files (
) | SAM/BAM Mapping Files (![]() )
)
This will open a dialog where you choose the reference sequences to be used as shown in figure 6.13.
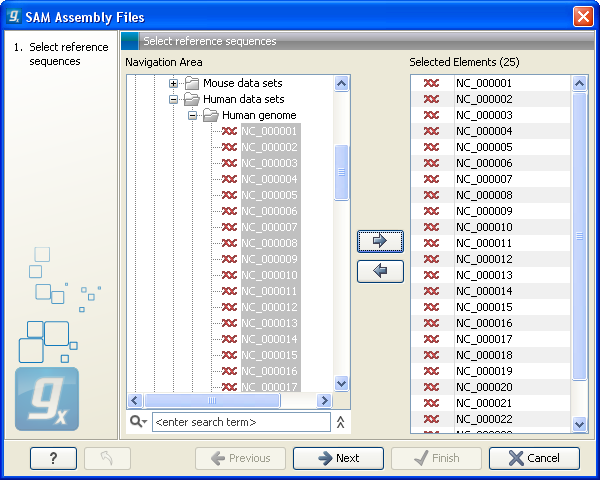
Figure 6.13: Defining reference sequences.
Select one or more reference sequence. Note that the name of your reference sequence has to match the reference name specified in the SAM/BAM file. Click Next.
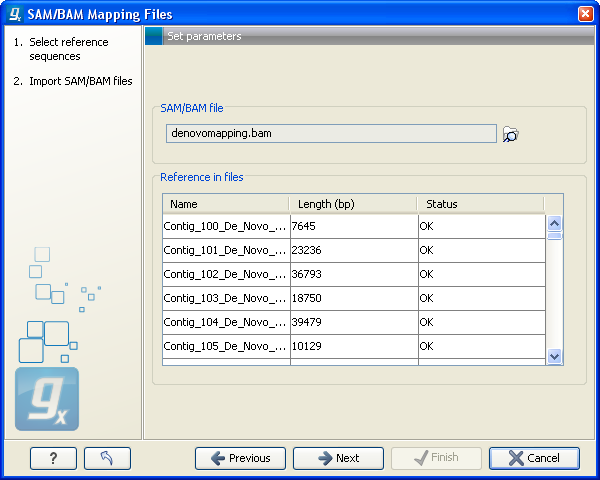
Figure 6.14: Selecting the SAM/BAM file containing all the read information.
In this dialog, select (![]() ) one or more SAM/BAM files as shown in figure 6.14.
) one or more SAM/BAM files as shown in figure 6.14.
In the panel below, all the reference sequences found in the SAM/BAM file will be listed included their lengths. In addition, it is indicated in the Status column whether they match the reference sequences selected from the Workbench. This can be used to double-check that the naming of the references are the same. (Note that reference sequences in a SAM/BAM file cannot contain spaces. If a reference sequence in the Workbench contains spaces, the space will be replaced with _ when comparing with the SAM/BAM file.). Figure6.15 shows an example where a reference sequence has not been provided (input missing) and one where the lengths of the reference sequences do not match (Length differs).
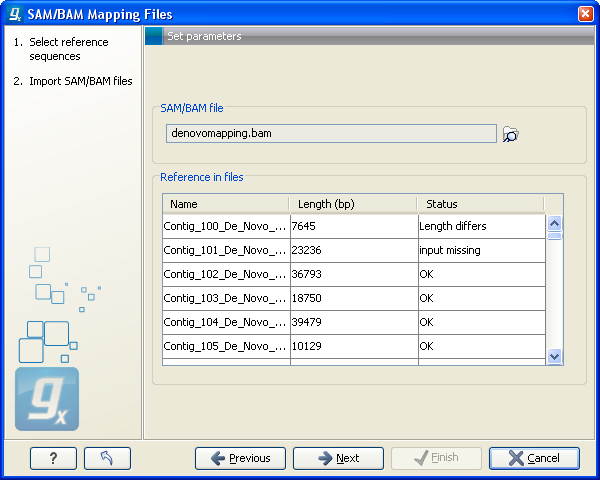
Figure 6.15: When there is inconsistency in the naming and sizes of reference sequences, this is shown in the dialog prior to import.
Click Next to adjust how to handle the results. We recommend choosing Save in order to save the results directly to a folder, since you probably want to save anyway before proceeding with your analysis.
Note that this import operation is very memory-consuming for large data sets.
