Extract sequences
It is possible to extract individual sequences from a sequence list in two ways. If the sequence list is opened in the tabular view, it is possible to drag (with the mouse) one or more sequences into the Navigation Area. This allows you to extract specific sequences from the entire list. Another option is to extract all sequences found in the list. This can also be done for:
- Alignments (
 )
)
- Contigs and read mappings (
 )
)
- Read mapping tables (
 )
)
- BLAST result (
 )
)
- BLAST overview tables (
 )
)
- RNA-Seq samples (
 )
)
- and of course sequence lists (
 )
)
To extract the sequences:
Toolbox | Classical Sequence Analysis (![]() ) | General Sequence Analysis (
) | General Sequence Analysis (![]() )| Extract Sequences (
)| Extract Sequences (![]() )
)
This will allow you to select the elements that you want to extract sequences from (see the list above). Clicking Next displays the dialog shown in 10.17.
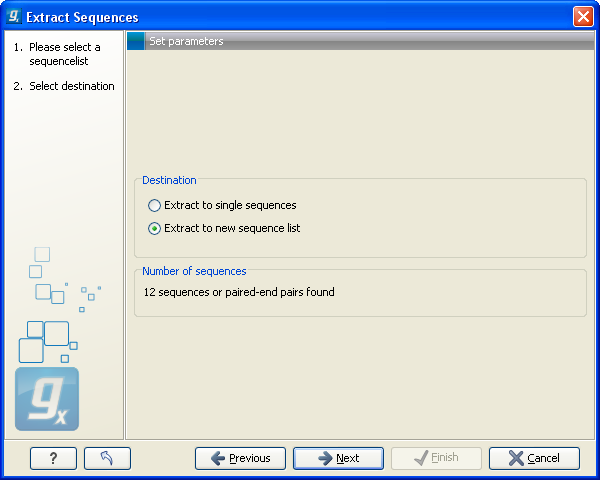
Figure 10.17: Choosing whether the extracted sequences should be placed in a new list or as single sequences.
Here you can choose whether the extracted sequences should be placed in a new list or extracted as single sequences. For sequence lists, only the last option makes sense, but for alignments, mappings and BLAST results, it would make sense to place the sequences in a list.
Below these options you can see the number of sequences that will be extracted.
Click Next if you wish to adjust how to handle the results. If not, click Finish.
