Assemble to reference sequence
This section describes how to assemble a number of
sequence reads into a contig using a reference sequence. A reference
sequence can be particularly helpful when the objective is to
characterize SNP variation in the data.
To start the assembly:
select sequences to assemble | Toolbox in the
Menu Bar | Molecular Biology Tools (![]() ) | Sequencing Data Analysis (
) | Sequencing Data Analysis (![]() )|
Assemble Sequences to Reference (
)|
Assemble Sequences to Reference (![]() )
)
This opens a dialog where you can alter your choice of sequences which you want to assemble. You can also add sequence lists.
Note! You can assemble a maximum of 2000 sequences at a time.
To assemble more sequences, please use the Map Reads to Reference (![]() ) under NGS Core Tools (
) under NGS Core Tools (![]() ) in the Toolbox.
) in the Toolbox.
When the sequences are selected, click Next, and you will see the dialog shown in figure 18.7
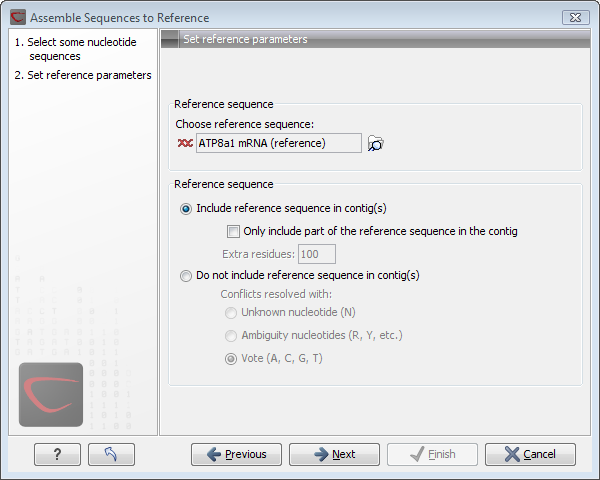
Figure 18.7: Setting assembly parameters when assembling to a reference sequence.
This dialog gives you the following options for assembling:
- Reference sequence. Click the Browse and select element icon (
 ) in order to select a sequence to use as reference.
) in order to select a sequence to use as reference.
- Include reference sequence in contig(s).
This will display a contig data-object with the reference sequence at the top
and the reads aligned below. This option is useful when comparing sequence reads to a closely related reference
sequence e.g. when sequencing for SNP characterization.
- Only include part of the reference sequence in the contig. If the aligned sequence reads only cover a small part of the reference sequence, it may not be desirable to include the whole reference sequence in the contig data-object. When selected, this option lets you specify how many residues from the reference sequence that should be kept on each side of the region spanned by sequencing reads by entering the number in the Extra residues field.
- Do not include reference sequence in contig(s).
This will produce a contig data-object without the reference sequence. The contig is created in the same way as when you make an
ordinary
assembly, but the reference sequence is omitted in the resulting contig. In the assembly process the reference sequence is only used as a scaffold for alignment.
This option is useful when performing assembly with a reference sequence that is not closely related to the sequencing reads.
- Conflicts resolved with. If there is a conflict, i.e. a position where there is disagreement about the residue (A, C, T or G), you can specify how the contig sequence should reflect this conflict:
- Unknown nucleotide (N). The contig will be assigned an 'N' character in all positions with conflicts.
- Ambiguity nucleotides (R, Y, etc.). The contig will display an ambiguity nucleotide reflecting the different nucleotides found in the reads. For an overview of ambiguity codes, see IUPAC codes for nucleotides.
- Vote (A, C, G, T). The conflict will be solved by counting instances of each nucleotide and then letting the majority decide the nucleotide in the contig. In case of equality, ACGT are given priority over one another in the stated order.
- Conflicts resolved with. If there is a conflict, i.e. a position where there is disagreement about the residue (A, C, T or G), you can specify how the contig sequence should reflect this conflict:
When the parameters have been adjusted, click Next, to see the dialog shown in figure 18.8
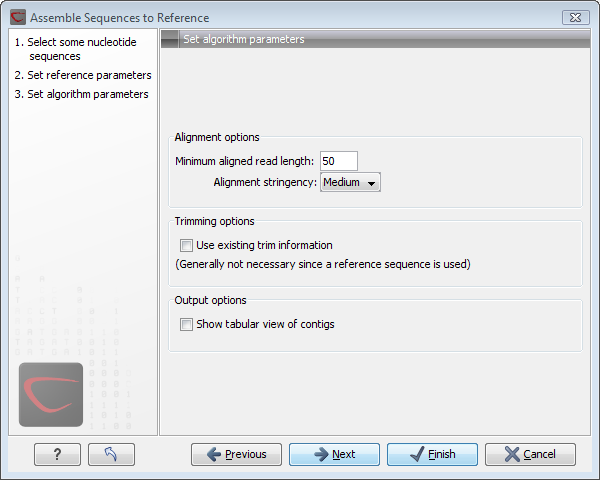
Figure 18.8: Different options for the output of the assembly.
In this dialog, you can specify more options:
- Minimum aligned read length. The minimum number of nucleotides in a read which must be successfully aligned to the contig. If this criteria is not met by a read, this is excluded from the assembly.
- Alignment stringency. Specifies the stringency of the scoring function used by the alignment step in the contig
assembly algorithm. A higher stringency level will tend to produce
contigs with less ambiguities but will also tend to omit more
sequencing reads and to generate more and shorter contigs. Three
stringency levels can be set:
- Low.
- Medium.
- High.
- Use existing trim information. When using a reference sequence, trimming is generally not necessary, but if you wish to use trimming you can check this box. It requires that the sequence reads have been trimmed beforehand (see Trim sequences for more information about trimming).
- Show tabular view of contigs. A contig can be shown both in a graphical as well as a tabular view.
If you select this option, a tabular view of the contig will also be
opened (Even if you do not select this option, you can show the
tabular view of the contig later on by clicking Show (
 )
and selecting Table (
)
and selecting Table ( ).) For more information about the
tabular view of contigs, see Assembly variance
table.
).) For more information about the
tabular view of contigs, see Assembly variance
table.
Click Next if you wish to adjust how to handle the results. If not, click Finish. This will start the assembly process. See View and edit contigs on how to use the resulting contigs.
