Running de novo assembly and read mapping in batch
De novo assembly and read mapping are special in batch mode because they usually have the option of assigning individual mapping parameters to each input file. When running in batch mode this is not possible. Instead, you can change the default parameters used for long and short reads, respectively. You can also set the paired distance for paired data.
Note that this means that you cannot use a combination of paired-end and mate-pair data for batching.
Figure 8.4 shows the parameter dialog when running read mapping in batch.
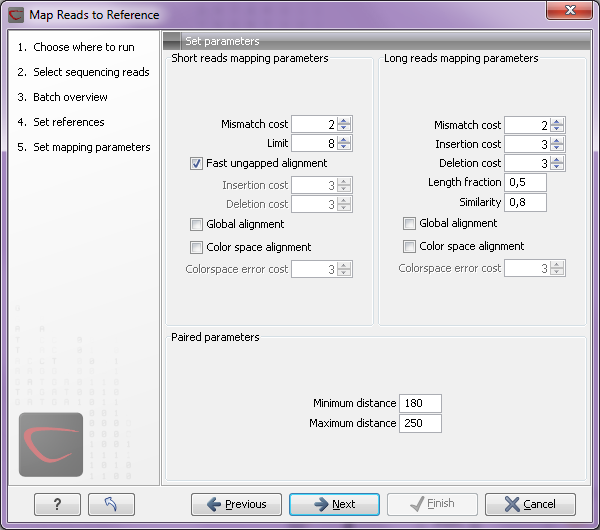
Figure 8.4: Read mapping parameters in batch.
Note that you can only specify one setting for all short reads, and one setting for all long reads. When the analysis is run, the reads are automatically categorized as either long or short, and the parameters specified in the dialog are applied. The same goes for all reads that are imported as paired where the minimum and maximum distances are applied.
