Join alignments
CLC Genomics Workbench can join several alignments into one. This feature can for example be used to construct "supergenes" for phylogenetic inference by joining alignments of several disjoint genes into one spliced alignment. Note, that when alignments are joined, all their annotations are carried over to the new spliced alignment.
Alignments can be joined by:
select alignments to join | Toolbox in the Menu
Bar | Classical Sequence Analysis (![]() ) | Alignments and Trees (
) | Alignments and Trees (![]() )|
Join Alignments (
)|
Join Alignments (![]() )
)
or select alignments to join | right-click
either selected alignment | Toolbox | Classical Sequence Analysis (![]() ) | Alignments and Trees (
) | Alignments and Trees (![]() )| Join Alignments
(
)| Join Alignments
(![]() )
)
This opens the dialog shown in figure 20.10.
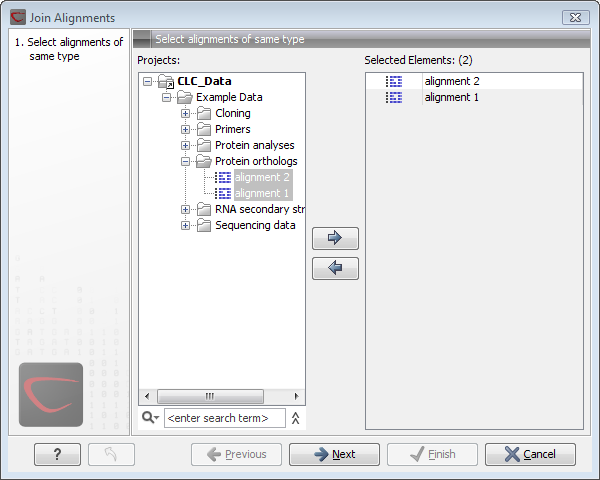
Figure 20.10: Selecting two alignments to be joined.
If you have selected some alignments before choosing the Toolbox action, they are now listed in the Selected Elements window of the dialog. Use the arrows to add or remove alignments from the selected elements. Click Next opens the dialog shown in figure 20.11.
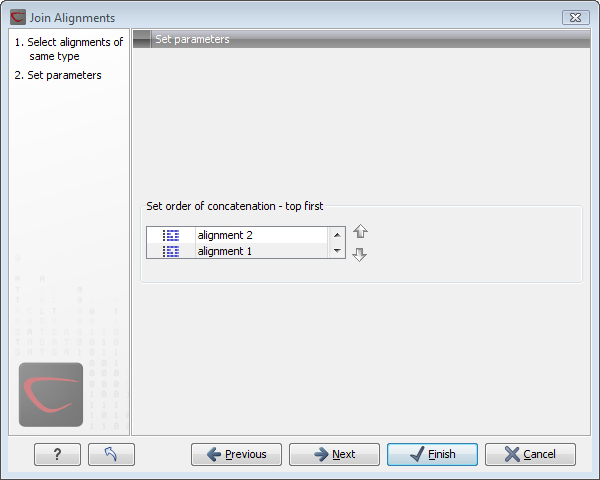
Figure 20.11: Selecting order of concatenation.
To adjust the order of concatenation, click the name of one of the alignments, and move it up or down using the arrow buttons.
Click Next if you wish to adjust how to handle the results. If not, click Finish. The result is seen in figure 20.12.
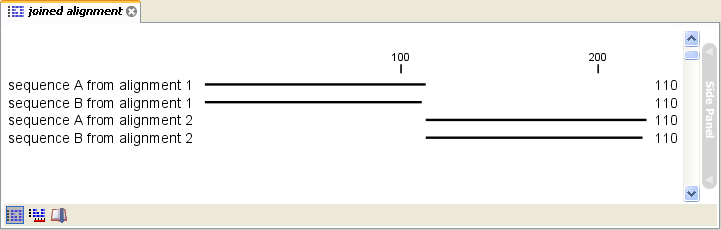
Figure 20.12: The joining of the alignments result in one alignment containing rows of sequences
corresponding to the number of uniquely named sequences in the joined alignments.
Subsections
