Finding annotations on the genome
In the Side Panel under Find, there is a search field that will take you to the annotation that you are looking for. There is a list of tracks that allow you to restrict the search to a particular track (e.g. a gene track).
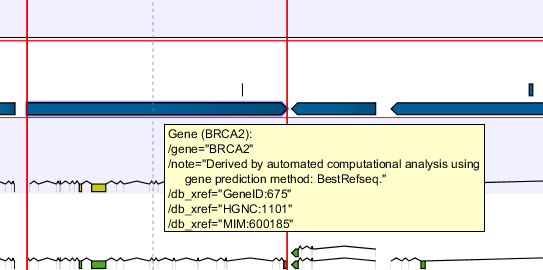
In the search field you can enter text to be found in the annotation track. As an example, consider the gene shown in figure 24.8
If you wanted to locate this gene, you could enter any of the following in the search field:
- BRCA2
- This would match the annotation name exactly.
- BRCA*
- This would match the annotation name as well as other genes with a text starting with BRCA (e.g. the BRCA1 gene).
- *RCA2
- This would match the annotation name as well as other genes with a text ending with RCA2 (e.g. the SMARCA2 gene).
- 600185
- This would match the
db_xrefqualifier for the OMIM database. All the text shown for the annotation in figure 24.8 can be searched this way, both as exact matches and with the * before or after the search term.
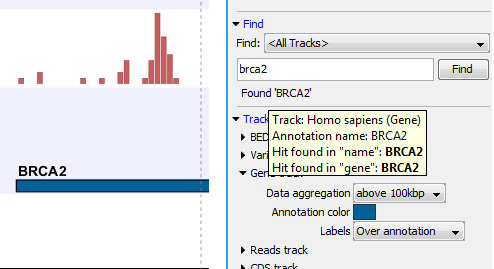
Figure 24.9: The BRCA2 gene found.
The search will run through the genome from the beginning of the chromosome currently shown, and it will stop when it finds the first hit. Press Find again to find the next hit. Once the whole genome has been traversed, the status will inform you that you have searched the whole genome, and you can click the Find button to start the search again.
Please note that you can also use the table view of an annotation track to do more advanced queries of the data (see Showing a track in a table).