Filter against known variants
Comparison with known variants from previous experiments or variant databases is a key concept when working with
resequencing data. The CLC Genomics Workbench provides two tools for facilitating this
task: one for annotating your experimental variants with information from
known variants (e.g. adding information about phenotypes like cancer
associated with a certain variant allele), and one for filtering your
experimental variants based on this information (e.g. for removing common
variants). The first tool is explained in Annotating known variants, while this section explains the latter.
In order to do this, you will have to import or download a file that is recognized as a variant file (see Import tracks from file and Download reference genome). 26.2.
This section will use the filter tool as an example, since the core of the tools are the same:
Toolbox | Resequencing (![]() ) | Annotate and Filter | Filter against Known Variants
) | Annotate and Filter | Filter against Known Variants
This opens a dialog where you can select a variant track (![]() ) with experimental data that should be filtered.
) with experimental data that should be filtered.
Clicking Next will display the dialog shown in figure 26.20
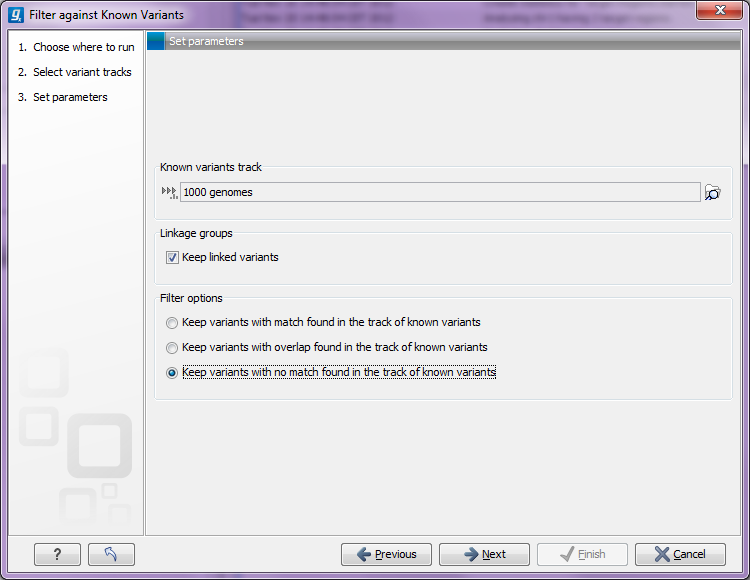
Figure 26.20: Specifying a variant track to filter against.
Select (![]() ) one or more tracks of known variants to compare against. The tool will then compare
each of the variants provided in the input track to see if it is reported in the track of known variants. There are three modes of filtering:
) one or more tracks of known variants to compare against. The tool will then compare
each of the variants provided in the input track to see if it is reported in the track of known variants. There are three modes of filtering:
- Keep variants found among known variants
- This will filter away all variants that are not found in the track of known variants. This mode can be useful for filtering against tracks with known disease-causing mutations (e.g. COSMIC), where the result will only include the variants that match the known mutations. For SNVs, the criteria for matching are simple: the variant position and allele both have to be identical in the input and the known variants track. For insertions and deletions, it is taken into account that they cannot always be placed unambiguously. As an example,
AA->Acan be a deletion of either the first or the second A, and both will be recognized as a match. For each variant found, the result track will include information from the known variant. - Keep variants overlapping with known variants
- The first mode is based on exact matching of the variants. This means that if the allele is reported differently in the set of known variants, it will not be identified as a known variant. This is typically not the case with isolated SNVs, but for more complex variants it can be a problem. Instead of requiring a strict match, this mode will keep variants that overlap with a variant in the set of known variants. This is a more conservative approach and will allow you to inspect the annotations on the variants instead of removing them when they do not match. For each variant, the result track will include information about overlapping or strictly matched variants to allow for more detailed exploration.
- Keep variants not found among known variants
- This mode can be used for filtering away common variants if they are not of interest. For example, you can download a variant track from 1000 genomes and use that for filtering away common variants. This mode is based on exact match. If you wish to filter based on overlap, please use the Filter against overlapping annotations tool.
The option to Keep linked variants comes into play for variants that are linked (see Linking adjacent variants in linkage groups). As an example, you may have a variant like this AC->GT. This is reported in the variant track as two separate variants in the same linkage group. If just one of the variants are found among the known variants, they will both be retained if the option to keep linked variants is checked. If the option is unchecked, it means that the linkage group in this situation will be broken and one of the variants will be removed.
Footnotes
- ...sec:downloadreferencegenome). 26.2
- Please note that there is also a plug-in for annotating with data from HGMD and other databases via Biobase Genome Trax: http://www.clcbio.com/clc-plugin/biobase-genome-trax/
