Find Resistance with ShortBRED
This tool allows you to detect and quantify the presence of antibiotic resistance (AR) marker genes that are represented by a database of peptide marker sequences. The tool first checks for exact matches against this database, and then runs DIAMOND. The Find Resistance with ShortBRED tool works similarly to the quantify step of ShortBRED, a public bioinformatics pipeline and resource. The tool is based on the ShortBRED-Quantify tool [Kaminski et al., 2015].
Find Resistance with ShortBRED quantifies the presence of Antibiotic Resistance (AR) marker genes in a sample of NGS short reads. It is possible to output a sequence list containing all the input reads which contained a marker (each read in the output is annotated with metadata describing the properties of the marker detected in the read).
Antibiotic Resistance marker databases for use with the tool can be downloaded using Download Resistance Database (Download Resistance Database).
To start the tool, go to:
Tools | Microbial Genomics Module (![]() ) | Drug Resistance Analysis (
) | Drug Resistance Analysis (![]() ) | Find Resistance with ShortBRED (
) | Find Resistance with ShortBRED (![]() ).
).
The tool accepts a nucleotide sequence or sequence list as input.
In the next dialog (figure 13.5), several parameters are available:
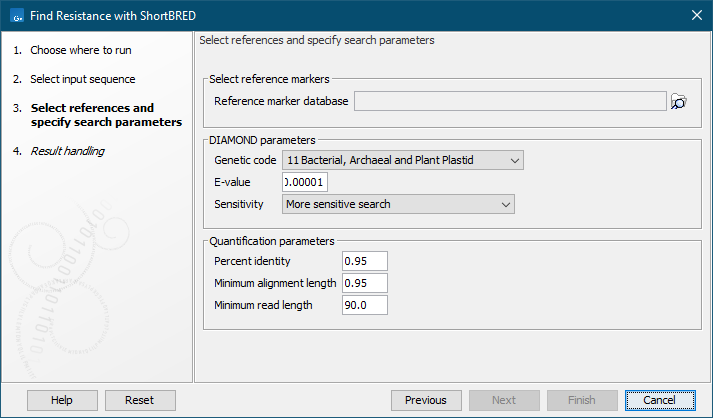
Figure 13.5: References and search parameters.
- Reference marker database: select the antimicrobial resistance marker database.
- Genetic code: select the genetic code to use when translating the nucleotide sequences to proteins.
- E-value: specify an expectation value to use as threshold for qualifying hits with DIAMOND. Note that exact matches to a peptide in the antimicrobial resistance marker database will always be reported, even if these would otherwise have an E-value larger than this threshold.
- Sensitivity: select DIAMOND sensitivity:
- Faster search: The fastest search
- Fast search: Designed for finding hits of >90% identity
- Standard search: Designed for finding hits of >60% identity
- Mid-sensitive search: More sensitive than standard search and faster than sensitive search.
- Sensitive search: Designed for finding hits of >40% identity
- More sensitive search: Designed for finding hits of >40% identity with some motif masking disabled
- Very sensitive search: Designed for finding hits of 40% identity
- Most sensitive search: The most sensitive search
- Percent identity: defines a minimum threshold for the percent identity of an alignment. This value is used by Find Resistance with ShortBRED to determine whether a hit found by DIAMOND is sufficiently good to be validated as a true hit (equivalent to the parameter "-id" of the ShortBRED-Quantify tool).
- Minimum alignment length: the minimum length of the DIAMOND alignment. This value is used by Find Resistance with ShortBRED to determine whether a hit found by DIAMOND is sufficiently good to be validated as a true hit (equivalent to the parameter "-pctlength" of the ShortBRED-Quantify tool - see citation at the beginning of this section).
- Minimum read length: the minimum read length. This value is used by Find Resistance with ShortBRED to determine whether a read is long enough to be processed by Find Resistance with ShortBRED (equivalent to the parameter "-minreadBP" of the ShortBRED-Quantify tool).
The Find Resistance with ShortBRED tool will output a resistance abundance table, a result summary table, an optional report, and an optional sequence list.
The result summary table provides an overview of all the resistance phenotypes in the applied database and reports the number of identified resistance genes and markers, and number of reads assigned to each phenotype.
The optional report output contains general information about the input sample, the marker database used and a short result summary.
The optional sequence list output contains all reads from the input sample which were found to contain one of the AR marker sequences. Each read in the sequence list is annotated with metadata describing the properties of the marker detected in the read.
Subsections
