Beta diversity measures
The following beta diversity measures are available:
- Bray-Curtis:
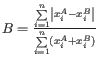
- Jaccard:
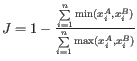
- Euclidean:
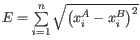
The following distances are also available:
- Unweighted UniFrac:
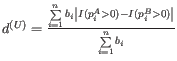
- Weighted UniFrac:
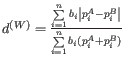
- Weighted UniFrac not normalized:
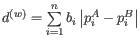
- D_0 UniFrac: The generalized UniFrac distance
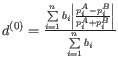
- D_0.5 UniFrac: The generalized UniFrac distance
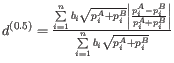
The unweighted UniFrac distance gives comparatively more importance to rare lineages, while the weighted UniFrac distance gives more important to abundant lineages. The generalized UniFrac distance ![]() offers a robust tradeoff [Chen et al., 2012].
offers a robust tradeoff [Chen et al., 2012].
