MLST Scheme Visualization and Management
An MLST Scheme contains information about:
- The loci that define the regions of interest.
- For each locus, a list of known alleles.
- A list of sequence types, where each sequence type is described by the alleles present at each locus (the profile of the sequence type).
The MLST Scheme has several views. Switching between the views of the scheme is done by clicking the buttons in the lower-left corner of the view.
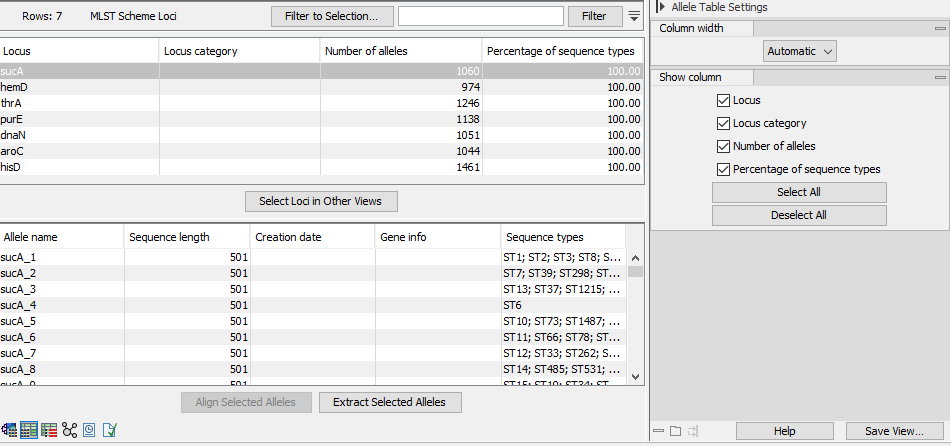
Figure 10.1: The Allele Table view.
The Allele Table view (figure 10.1) has an upper table that lists the loci in the scheme. The table contains the following columns:
- Locus. The name of the locus.
- Locus category. Shows any virulence or resistance-gene related annotations.
- Number of alleles. The total number of alleles for this locus.
- Percentage of sequence types. Shows how many of the sequence types have an allele in the given locus. For a strict core genome MLST (scgMLST) scheme, all of the sequence types contain all loci.
The lower table lists the alleles for the selected loci. It has the following columns:
- Allele name. The name of the allele.
- Sequence length. Length in nucleotides.
- Creation date. When the allele was added (if scheme was created using Create MLST Scheme).
- Gene info. AMR or virulence related information.
- Sequence types. The sequence types that contain this allele.
- Percentage. Shows how many sequence types have this specific allele.
It is possible to Align Selected Alleles, which creates a new multiple sequence alignment view or to Extract Selected Alleles, which creates a sequence list with the alleles.
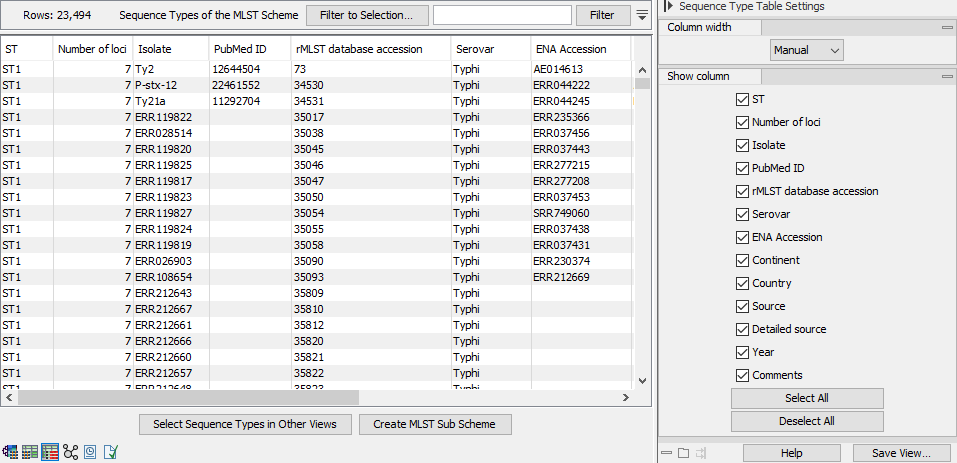
Figure 10.2: The Sequence Type Table view.
The Sequence Type Table view (figure 10.2) shows the sequence types in the scheme. It always contains the following columns:
- ST. The name of the sequence type.
- Number of loci. The number of loci, that are defined for this sequence type. scgMLST schemes and classic 7-gene schemes will have the same number of loci for all sequence types.
If the scheme contains any metadata information, columns representing each metadata category may be present as well.
At the bottom of the view, two buttons make it possible to Select Sequence Types in Other Views and to Create MLST Sub Scheme:
- Select Sequence Types in Other Views. Selected sequence types will be selected in an opened and linked Minimum Spanning Tree or Sequence Type Table view, either from the same MLST Scheme or an MLST Comparison made from the same MLST Scheme. Selecting one or more sequence types in one of the views, followed by clicking the button, will result in the same sequence types being selected in the other view.
- Create MLST Sub Scheme. This makes it possible to create a new scheme based on a selection of sequence types in the scheme.
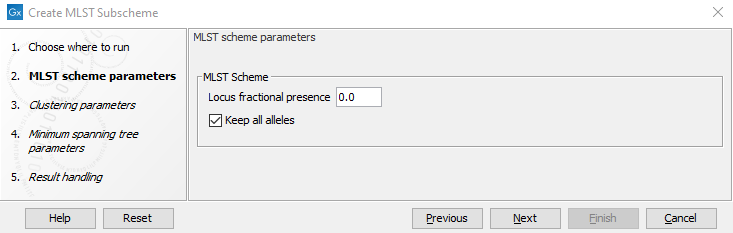
Figure 10.3: The Create MLST Subscheme options.
Create MLST Sub Scheme has the same options as the other scheme creation tools, except for some additional options for pruning the scheme (figure 10.3):
- Locus fractional presence. The fraction of sequence types required to have an allele specified for a given locus before the locus is added to the new scheme. For instance, a value of 0.95 would mean that the resulting scheme only contains loci present in at least 95% of the selected sequence types (a loose core genome scheme).
- Keep all alleles. If this option is deselected, only alleles that are part of at least one sequence type are retained. Alleles from discarded loci will always be removed.
The MLST Scheme also has a Minimum Spanning Tree view, which is the topic of the next section.
