The MLST Typing Result element
The MLST Typing Result element contains several views. Switching between the views of the scheme is done by clicking the buttons at the lower-left corner of the view. The number of Sequence Types shown is limited to 100 or the number of Sequence Types in the scheme, depending on which is lower.
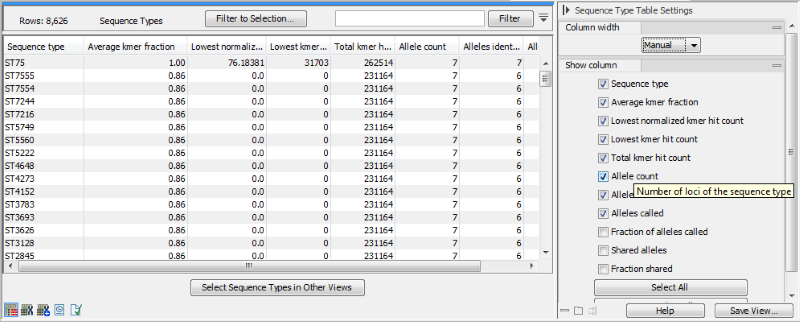
Figure 10.15: The sequence type table for a MLST Typing Result.
The sequence type table is a tabular view with information about how well the sample matched the sequence types in the scheme (figure 10.15). It contains the following columns:
- Sequence type: name of the sequence type
- Average kmer fraction: for all alleles in a given sequence type, we calculate how many of the allele's kmers were detected. This number is the average fraction of the number of kmers detected in all these alleles.
- Lowest normalized kmer hit count: Normalized kmer hit count for the allele with the lowest normalized hit count of the sequence type.
- Lowest kmer hit count: Number of kmer hits for the allele with the fewest hits of the sequence type.
- Total kmer hit count: sum of kmer hits for all alleles of the sequence type
- Allele count: the total number of alleles in the sequence type.
- Alleles identified: the number of alleles in the sequence type where all kmers of the allele were found in the sample.
- Alleles called: the number of alleles in the sequence type with at least one kmer found in the sample.
- Fraction of alleles called: the ratio between alleles called and the allele count for the sequence type.
- Shared alleles: Number of alleles shared with the best scoring sequence type
- Fraction shared: Fraction of alleles shared with the best scoring sequence type
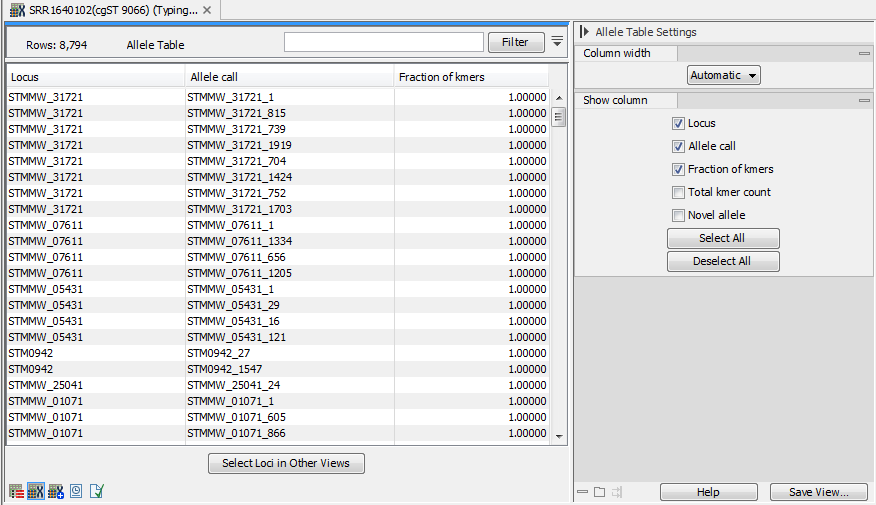
Figure 10.16: The allele table for a MLST Typing Result.
The allele table (figure 10.16) contains information about the alleles that were identified in the sample. It contains the following columns:
- Locus: the name of the locus.
- Allele call: the allele that was identified for that locus. Only the best allele is reported, but if multiple alleles are tied for the first place, they will all be reported.
- Fraction of kmers: the fraction of kmers of the allele that were found in the sample.
- Total kmer count: total number of kmer hits for the allele.
- Novel allele: contains the string 'Novel' for novel alleles, otherwise this field is left blank.
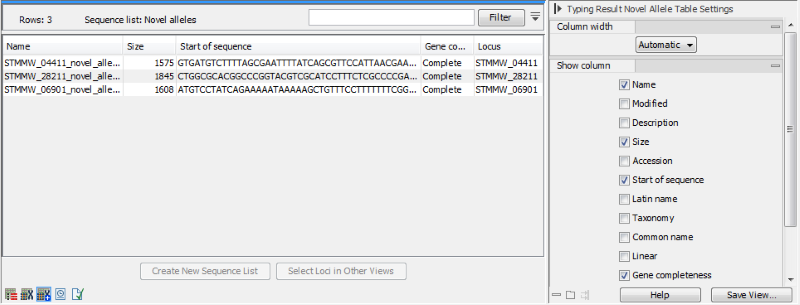
Figure 10.17: The novel allele view for a MLST Typing Result.
The novel allele view (figure 10.17) contains the novel alleles that were detected (if searching for novel alleles was enabled during the typing).
It is a sequence list, and it is possible to extract the complete sequences using the Create New Sequence List button.
The Gene completeness column is the only non-standard sequence list column: if a novel allele starts with a start codon and ends with a stop codon it is considered complete. Note that all novel alleles found with a scheme without a translation code will be incomplete.
