Import MLST Scheme
To run the Import MLST Scheme tool, go to:
Tools | Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | MLST Typing (
) | MLST Typing (![]() ) | Import MLST Scheme (
) | Import MLST Scheme (![]() )
)
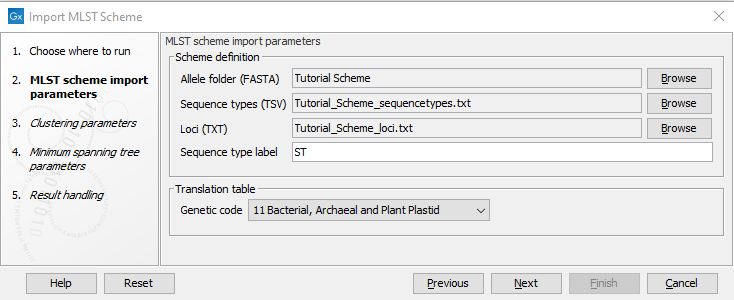
Figure 14.12: The MLST Scheme import parameters.
The Name parameter allows you to set the output name for the MLST scheme.
The Alleles must contain a set of FASTA files, one for each locus, or a single compressed ZIP archive containing all these files. The files must have one of the following extensions to be recognized: "fa", "fas", "fsa", "fasta", "tfa". The name of the allele must be the locus name and allele name separated by an underscore, like in this example:
>pheS_1 AGAGAAAAGAACGATACTTTCTATATGGCCCGTGATAATCAAGGCAAGCGTGTTGTCTTA >pheS_2 AGAGAAAAGAACGATACTTTCTATATGGCCCGTGATAATCAAGGCAAGCGTGTTGTCTTA
The Sequence types file must be a tab-separated file listing a sequence type and its alleles in the following format:
ST pheS glyA fumC mdh sucA dnaN atpA clonal_complex 1 30 1 1 1 1 1 1 6 3 6 8 7 3 4 3 1 4 7 9 8 3 5 2 1 6 47 3 10 4 7 2 2It is possible to add arbitrary metadata as additional columns after the loci columns (e.g. the 'clonal_complex' column above). If multiple isolates share the same sequence type, but have different metadata, it is possible to add multiple lines with the same sequence type name and allele ids, but with different metadata entries.
The Loci file must be a tab-separated file listing a locus name and its corresponding metadata. For this file the only recognized headers are "Locus", "Known name", "Type name", "Locus type" where the name of the locus in the MLST scheme needs to match the name in the Locus column of the annotation file.
Locus Known name Type name Locus type locus5 FALSE Unknown ST1 fliR TRUE fli ST2 flgL TRUE flg ST3 hpaB TRUE hpa ST4
The Minimum Spanning Tree parameters are similar to the options for Download MLST Scheme tool (see Download MLST Scheme)
The genetic code specified will be used for novel allele detection to make sure each allele starts and ends with an initiation and stop codon, respectively. If "No code specified" is selected, these requirements will not be checked when searching for novel alleles, instead the aligned part of the existing alleles to the assembly is used to define the allelic length. Note that the latter is useful for 7-gene MLST schemes which generally use fractions of genes, but it is also sensitive towards unaligned ends and may return too short alleles in some cases.
