Mask Low-Complexity Regions
The Mask Low-Complexity Regions tool can be used to identify and mask repetitive regions in sequences. In some cases this can remove erroneous matches: for instance, when doing taxonomic profiling, a read with a highly repetitive sequence is likely to match a reference genome purely by chance.
The tool takes any sequence or sequence list as input (including reads and genomes). It will accept both nucleotide and protein sequence input.
To run the tool, go to
Tools | Utility Tools (![]() ) | Mask Low-Complexity Regions (
) | Mask Low-Complexity Regions (![]() )
)
The following general options are available (figure 20.1):
- Window size: The complexity is evaluated by moving a window along the sequences. This option sets the sliding window size.
- Window stride: The number of nucleotides by which the window is moved along the sequence. Increasing this value makes the tool faster, but slightly less accurate when detecting the edges of low-complexity regions.
- Low-complexity threshold: This measure is normalized such that a value of 0 corresponds to a trivial sequence (e.g. 'AAAAAAAA'), and 1 corresponds to a random sequence. Higher values mask more of the sequence. Notice, that the report contains sequence examples for different complexity thresholds.
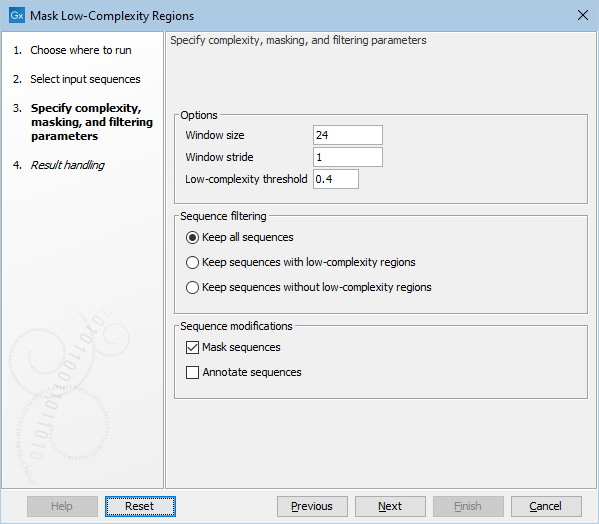
Figure 20.1: The Mask Low-Complexity Regions options.
The Sequence filtering options make it possible to specify whether some or all input sequences should be output:
- Keep all sequences. No filtering is performed.
- Keep sequences with low-complexity regions. Notice, that sequences which have already been masked in a previous run of the tool will not be kept.
- Keep sequences without low-complexity regions. Note, that this also keeps sequences where low complexity regions have already been masked in a previous run of the tool.
Finally, the Sequence modifications options determine how the output sequences are marked:
- Mask sequences: Low-complexity regions are masked with N's (or X's for proteins). Notice, that if a tested window already contains ambiguous symbols (e.g. from a previous run of the tool), it will not be masked.
- Annotate sequences: Low-complexity regions are marked with a sequence annotation. Notice, that if a tested window already contains ambiguous symbols (e.g. from a previous run of the tool), it will not be marked.
The tool optionally outputs a report with statistics on the detected regions. The report is described in details below:
Subsections
