Classify Whole Metagenome Data parameters
To run the Classify Whole Metagenome Data tool, go to
Tools | Microbial Genomics Module (![]() ) | Metagenomics (
) | Metagenomics (![]() ) | Taxonomic Analysis (
) | Taxonomic Analysis (![]() ) | Classify Whole Metagenome Data (
) | Classify Whole Metagenome Data (![]() )
)
In the first dialog, select the sequence list to analyze (figure 6.12).
Figure 6.12: Select a sequence list as input.
Click Next to select reference databases (figure 6.13):
- Reference index. Select the metagenome index. If your sample contains host reads, the index should include the host genome.
Curated reference databases and metagenome indexes are available from Download Curated Microbial Reference Database (Download Curated Microbial Reference Database). Alternatively, create your own index using Create Whole Metagenome Index (Create Whole Metagenome Index). - Host genome. Define the taxa of the host or background genome(s) that may be present in your sample, but that is not the target of your analysis. Reads assigned to the host genome are omitted from the abundance table output. As an example, to analyze the microbiome from human gut samples and exclude human reads from the result, specify the taxon "Homo sapiens" as host genome.
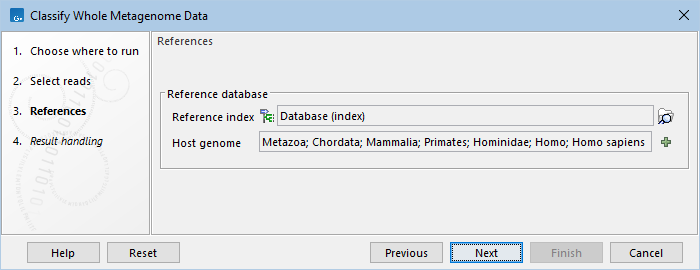
Figure 6.13: Select the reference index and, if relevant, define the host genome taxa.
