Add Typing Results to MLST Scheme
After typing an isolate, it is possible to add the information to the MLST Scheme. There are three different possibilities when adding a typing result:
- The typing result may have matched an existing sequence type completely. In this case, it is still possible and useful to add the typing result to the scheme, in order to add additional isolate metadata for the sequence type which may be from metadata annotations on the sequences or from a metadata table.
- The typing result may have matched existing alleles in the scheme but in a new combination not present in any of the existing sequence types. In this case, the typing result will simply be added as a new sequence type.
- The typing result may introduce new (novel) alleles to the scheme. In this case, both a new sequence type and one or more alleles are added to the scheme.
Add Typing Results to MLST Scheme is available from:
Tools | Microbial Genomics Module (![]() ) | Typing and Epidemiology (
) | Typing and Epidemiology (![]() ) | MLST Typing (
) | MLST Typing (![]() ) | Add Typing Results to MLST Scheme (
) | Add Typing Results to MLST Scheme (![]() )
)
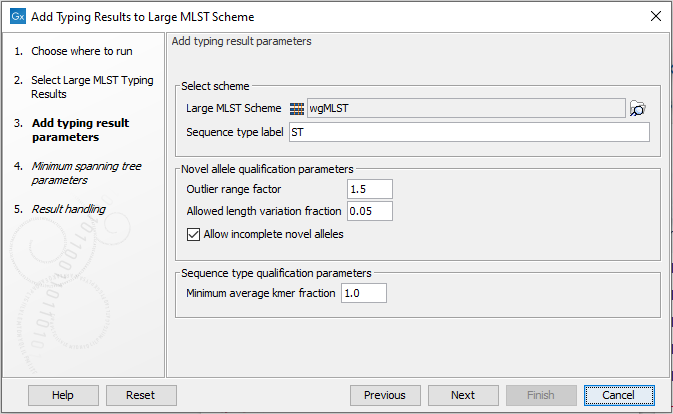
Figure 10.19: Add typing result parameters.
After selecting the MLST Typing result to add, the next step is to set up the Add typing result parameters (figure 10.19):
- MLST Scheme. The scheme that the typing results will be added to. Adding the typing results will not modify the original scheme, but create a new copy with the added types.
- Outlier range factor. The allele length outlier definition in terms of the interquartile range of the length distribution of alleles in a locus. Novel alleles outside this range will not be added to the scheme.
- Allowed length variation fraction. The allowed length variation of a novel allele with respect to the median length of alleles in a locus. This allows adding novel alleles with minor length variations irrespective of the outlier definition.
- Allow incomplete novel alleles. Whether only complete novel alleles (containing both start and stop codon) should be allowed. If incomplete novel alleles are not allowed, a sequence type with incomplete alleles for a locus will be added with missing alleles for that locus. Classic 7-gene schemes typically contain partial gene fragments, and in this case incomplete novel alleles should be allowed.
- Minimum average kmer fraction. If this value is larger than zero, a typing result will only be added if the isolate was sufficiently similar to an already existing sequence type in the scheme - or to put it differently - at least one of the sequence types in the MLST Typing Result must have an Average kmer fraction larger than this threshold. Note, that when typing against an empty scheme, this value must be set to zero, to allow for the sequence type to be added. This option is mostly useful when adding a large number of isolates in bulk without manually inspecting them.
After adding new sequence types it is necessary to recreate the Minimum Spanning Tree. The options are the same as described in the Download MLST section (Download MLST Scheme)
