Create Whole Metagenome Index
This tool will generate a whole metagenome index from a reference database. This index type is used by the Classify Whole Metagenome Data tool (Classify Whole Metagenome Data).
It is recommended to mask repetitive sequences in the reference database (host genome excluded) with Mask Low-Complexity Regions (Mask Low-Complexity Regions) prior to running Create Whole Metagenome Index.
To run the tool, go to:
Tools | Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Taxonomic Analysis (
) | Taxonomic Analysis (![]() ) | Create Whole Metagenome Index (
) | Create Whole Metagenome Index (![]() )
)
Select a sequence list containing the references and the potential host genome of interest.
All sequences must have a taxonomy attribute. Sequences and their associated taxonomy can be downloaded, for example, using Download Custom Microbial Reference Database (Download Custom Microbial Reference Database).
The output includes a whole metagenome index file and an optional report. The report provides a summary of the number of features at each taxonomic level, as well as information about the number of sequences and bases that were indexed (figure 16.7). "Number of genomes" is the number of unique assembly IDs. Sequences without assembly IDs are counted as a unique genome per sequence.
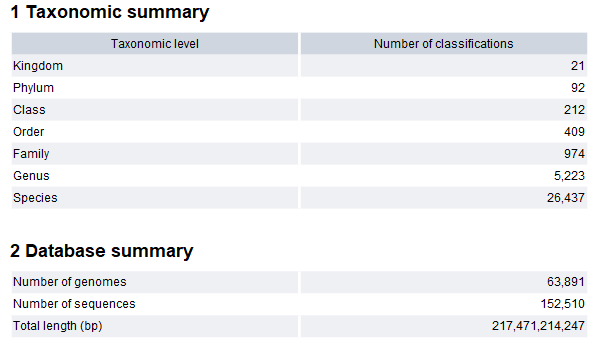
Figure 16.7: The Create Whole Metagenome Index report.
Depending on the size of the database, the tool may require a significant amount of free temporary disk space, see (System requirements).
The tool allows for creating indexes containing up to 65,535 taxonomic nodes, with each node corresponding to a taxonomic classification. For example, the index in figure 16.7 includes a total of 33,368 taxonomic classifications, or nodes.
