Clustering of features and samples
Hierarchical clustering clusters taxa by the similarity of their taxonomic profiles over the set of samples, and samples by the similarity of taxonomic composition over the set of features (taxa).Each clustering has a tree structure that is generated as follows:
- Letting each taxa or sample be a cluster.
- Calculating pairwise distances between all clusters
- Joining the two closest clusters into one new cluster.
- Iterating 2-3 times until there is only one cluster left (which contains all the taxa or samples).
In the resulting tree, the length of branches reflect the distance between clusters.
To create a heat map, run the Create Heat Map for Abundance Table tool, available from:
Tools | Microbial Genomics Module (![]() ) | Metagenomics (
) | Metagenomics (![]() ) | Abundance Analysis (
) | Abundance Analysis (![]() ) | Create Heat Map for Abundance Table (
) | Create Heat Map for Abundance Table (![]() )
)
Select an abundance table with two or more samples as input (e.g., a multi-sample OTU or merged abundance table) and click Next.
Specify a distance measure and a cluster linkage (figure 7.18). The distance measure specifies how distances between two taxa or samples should be calculated. The cluster linkage specifies how the distance between two clusters, each consisting of a number of taxa or samples, should be calculated. For information on how distances and clusters are calculated, see Normalization and clustering.
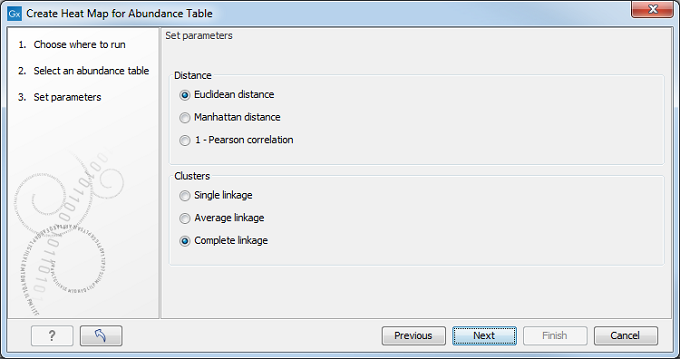
Figure 7.18: Select an abundance table.
After having selected the distance measure, set up the feature filtering options (figure 7.19).
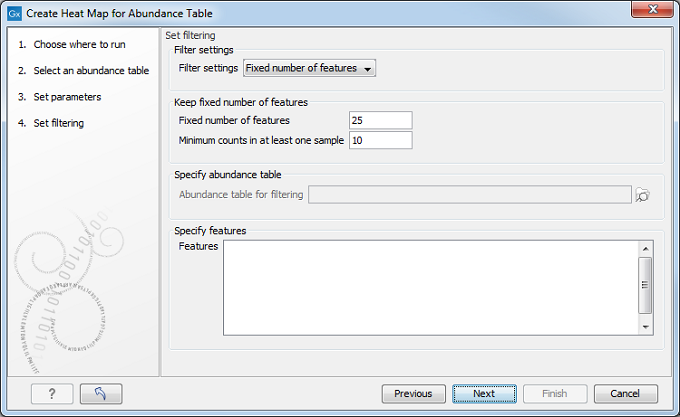
Figure 7.19: Set filtering options.
Genomes usually contain too many features to allow for a meaningful visualization. Clustering hundreds of thousands of features is also very time consuming. We therefore recommend to reduce the number of features before clustering, using the filter options available:
- No filtering: Keeps all features.
- Fixed number of features:
- Fixed number of features: The given number of features with the highest coefficient of variation (the ratio of the standard deviation to the mean) are kept.
- Minimum counts in at least one sample: Only features with more than this number of counts in at least one sample will be taken into account. Notice that the counts are raw, un-normalized values.
- Abundance table: Specify a subset of an abundance table in case you only want to display the heat map for that particular subset. Note that creating the heat map from the subset abundance table directly can not ensure proper normalization of the data, and it is therefore recommended to use the original abundance table as input and filter using this option.
- Specify features: Keeps a set of features, as specified by plain text, i.e., a list of feature names. Any white-space characters, as well as "," and ";" are accepted as separators.
