Add Metadata to Abundance Table
It is useful to have abundance tables decorated with sample metadata. This can be done by importing metadata and associating it with the reads before generating an abundance table. To learn more about how to create a metadata table, how to import a metadata table, or how to associate data elements with metadata, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Metadata.html.
If you did not have metadata associated with your reads prior to creating an abundance table, use Add Metadata to Abundance Table to add it retrospectively.
To run the Add Metadata to Abundance Table tool, go to:
Tools | Microbial Genomics Module (![]() ) | Metagenomics (
) | Metagenomics (![]() ) | Abundance Analysis (
) | Abundance Analysis (![]() ) | Add Metadata to Abundance Table (
) | Add Metadata to Abundance Table (![]() )
)
Choose an abundance table as input. In the next wizard window you can select a file describing the metadata on your local computer (figure 7.21).
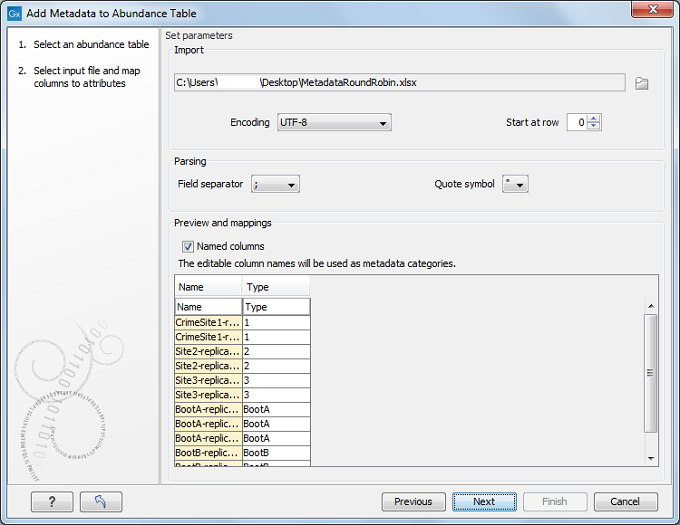
Figure 7.21: Setting up metadata parameters.
The metadata should be formatted in a tabular file format (*.xls, *.xlsx). The first row of the table should contain column headers. There should be one column called "Name" and the entries in this column should match the names of the reads selected for creating the abundance table. This column is used to match row in the table with samples present in the abundance table, so if the names do not match you will not be able to aggregate your data at all. There can be as many other columns as needed, and these information can be used as grouping variables to improve visualization of the results or to perform additional statistical analyses. If you wish to ignore a column without deleting it from your file, simply delete the text in the header row.
Note that when importing an Excel file, formulas will be imported as the formula text and not as the result of the calculation.
If you utilize formulas in the metadata file you want to import, you have to flatten the file before importing.
This can be done in a number of ways, for instance by exporting to a CSV file (and then importing that instead), or copying and using "Paste Special" in Excel:
Start by selecting everything, copy the selection to the clipboard and then execute "Paste Special".
On Windows "Paste Special" can be executed by holding Ctrl and Alt and then pressing V.
On a Mac "Paste Special" can be executed by holding Ctrl and ![]() and pressing V.
Once the "Paste Special" dialog appears, select "Values" under "Paste" and finally click OK.
and pressing V.
Once the "Paste Special" dialog appears, select "Values" under "Paste" and finally click OK.
