Spoligotype Mycobacterium Tuberculosis parameters
To run the Spoligotype Mycobacterium Tuberculosis tool, go to:
Tools | Microbial Genomics Module (![]() ) | Typing and Epidemiology (
) | Typing and Epidemiology (![]() ) | Spoligotype Mycobacterium Tuberculosis (
) | Spoligotype Mycobacterium Tuberculosis (![]() )
)
The tool takes sequence lists as input.
The tool will accept any reads, but as spoligotyping targets genomic regions in M. tuberculosis, you are unlikely to get meaningful results if the reads are not from the M. tuberculosis complex or its closely related strains. To avoid matching in off-target sequences, it is best practice to trim reads for adapters, quality, etc. In non-isolate samples it can also be beneficial to extract M. tuberculosis reads first e.g., by using Taxonomic Profiling before typing.
In the wizard, minimum coverage thresholds can be set (figure 11.1):
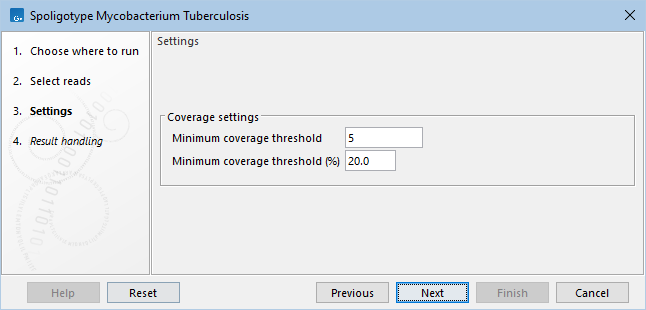
Figure 11.1: Spoligotype Mycobacterium Tuberculosis parameters.
- Minimum coverage threshold (count). The minimum number of times a spacer must be found for it to be considered present in the sample.
- Minimum coverage threshold (%). The minimum percent of times, relative to the maximum across all spacers, a spacer must be found for it to be considered present in the sample.
Both thresholds must be met for a spacer to be present. To apply only a count threshold, set the percentage to 0. Conversely, to apply only a percentage threshold, set the count to 1.
