Taxonomic Profiling output
Clicking Next will allow you to specify the output as shown in figure 6.21.
Figure 6.21: Specify the output.
The following outputs are available:
- Collect reference database reads. Creates a sequence list for each input with reads that were assigned to the reference database.
- Collect host genome reads. Creates a sequence list for each input with reads that were assigned to the host genome.
- Collect unmapped reads. Creates a sequence list for each input with unassigned reads.
- Create report. Creates a summary report.
Sequence list output
The sequence lists with reads that were assigned to the reference database and host genome contain the following annotations:
- Mapping Quality Score. Reads with quality score <10 will have been assigned to a higher taxonomy level.
- Mapping Score. The score for the read alignment (see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Mapping_parameters.html).
In addition, the list of reference database reads contains the Taxonomy annotation with the full taxonomy of the taxon to which the read was assigned.
Taxonomic Profiling report
The Taxonomic Profiling report (figure 6.22) contains information about the taxonomic profiling run and databases used.
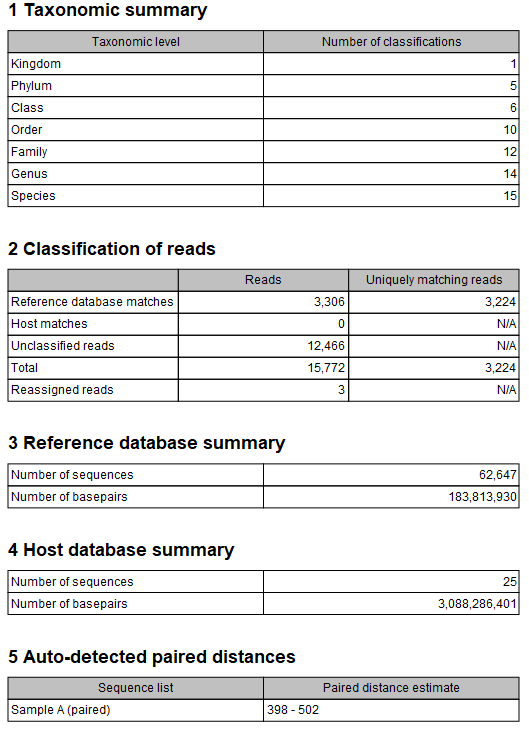
Figure 6.22: The Taxonomic Profiling report.
- Taxonomic summary. The number of detected taxa for each taxonomic level.
- Classification of reads. Information about the number of reads that were assigned to reference and host databases, or left unclassified. For database matches, both the total number of reads and the number of uniquely matched reads are provided.
- Reference database summary. Information about the reference database. Reads that map across Kingdoms will count as a reference database match, but will not contribute to values in the abundance table as no meaningful taxon can be assigned.
- Host database summary. Information about the host genome.
- Auto-detect paired distances. The calculated paired distance range (provided when the corresponding option was applied).
