Create MLST Scheme with Sequence Types
The Create MLST Scheme with Sequence Types workflow creates a MLST scheme from references and adds sequence types by typing references and adding the results to the scheme.To run the Create MLST Scheme with Sequence Types template workflow, go to
Workflows | Template Workflows (![]() ) | Microbial Workflows (
) | Microbial Workflows (![]() ) | Typing and Epidemiology (
) | Typing and Epidemiology (![]() ) | Create MLST Scheme with Sequence Types (
) | Create MLST Scheme with Sequence Types (![]() )
)
You can select one or more assemblies as input (figure 2.9). At least one of the assemblies must be annotated with CDS regions.

Figure 2.9: Select the high-quality references serving as the basis for the scheme
In "Create MLST scheme" dialog (figure 2.10), the settings for the scheme creation can be viewed and changed.
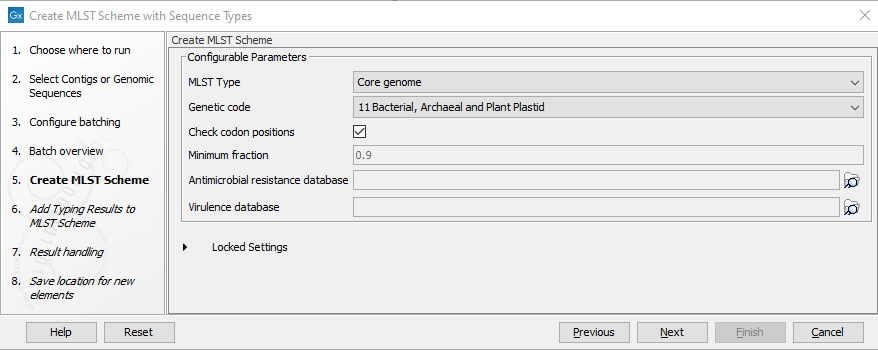
Figure 2.10: Parameters for creating the initial scheme
The parameters that can be set are:
- MLST Type: specifies the fraction of assemblies a locus must be present in to be included in the scheme. Options are: Core genome (corresponding to a fraction of 0.9), Whole genome (corresponding to a fraction of 0.1) or custom fraction.
- Genetic code: specifies the genetic code matching the input assemblies for a codon check.
- Check codon positions: if enabled, loci failing the specified codon check will not appear in the scheme. This should be disabled when working with organisms containing spliced genes.
- Minimum fraction: specifies the required fraction if custom fraction was selected in MLST Type.
- Antimicrobial resistance database: optional setting for specifying an antimicrobial resistance database to use for annotating loci in the scheme.
- Virulence database: optional setting for specifying a virulence database to use for annotating loci in the scheme.
In "Add Typing Results to MLST scheme" dialog (figure 2.11), sequence types will be added to the scheme. In addition, the following parameters can be specified:
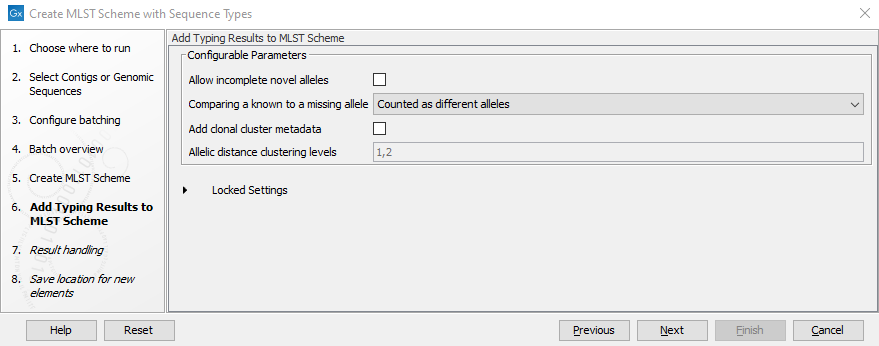
Figure 2.11: Add Typing Results settings
- Allow incomplete novel alleles: whether only complete novel alleles (containing both start and stop codon) should be allowed. If incomplete novel alleles are not allowed, a sequence type with incomplete alleles for a locus will be added with missing alleles for that locus. If Check codon positions has been disabled (see figure 2.10), all alleles will be incomplete and consequently it will be necessary to allow adding incomplete alleles.
- Comparing a known to a missing allele: how to treat missing alleles when comparing a locus for a pair of sequence types.
- Add clonal cluster metadata: if selected, clonal cluster data will be added as metadata.
- Allele distance clustering levels: if clonal cluster data is added, specifies the allelic distance thresholds for adding clustering information.
In the Result handling window, pressing the button Preview All Parameters allows you to preview - but not change - all parameters. Saving the output will generate the files shown in (figure 2.12) and optionally, a workflow result metadata table.
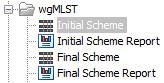
Figure 2.12: The output from Create MLST Scheme with Sequence Types
- Initial Scheme Report: the report from Create MLST Scheme tool.
- Initial Scheme: an empty scheme containing only loci.
- Final Scheme Report: the report from Add Typing Results to MLST Scheme tool.
- Final Scheme: the complete MLST Scheme containing loci and sequence types.
For more information on the tools and MLST schemes, see MLST Scheme Tools.
